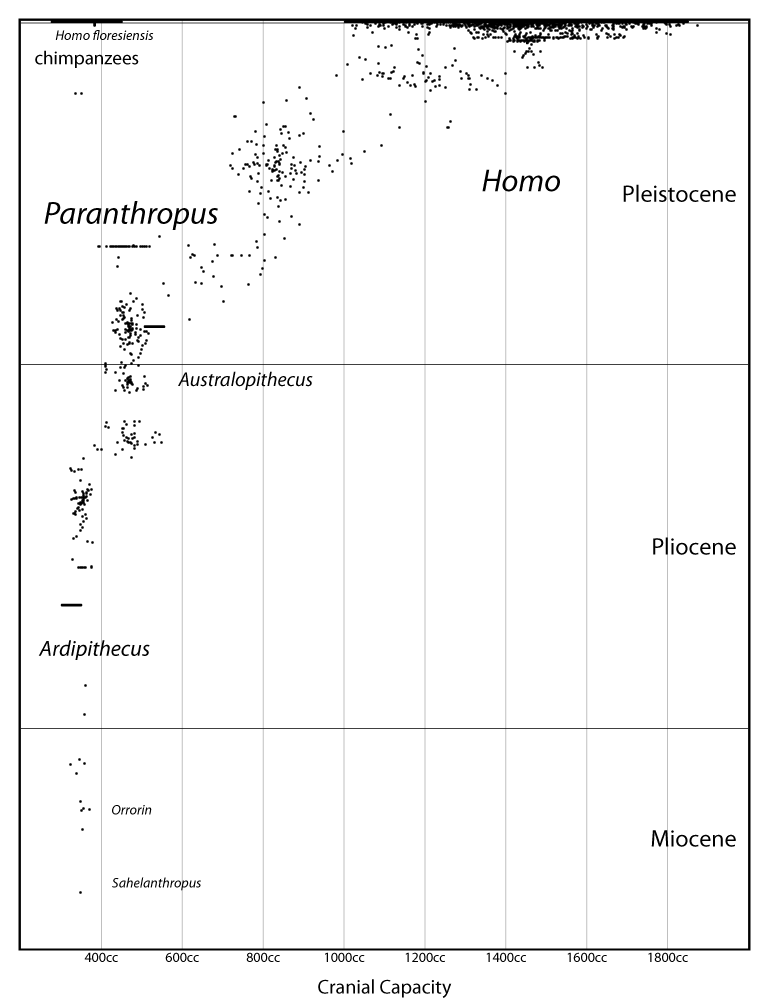
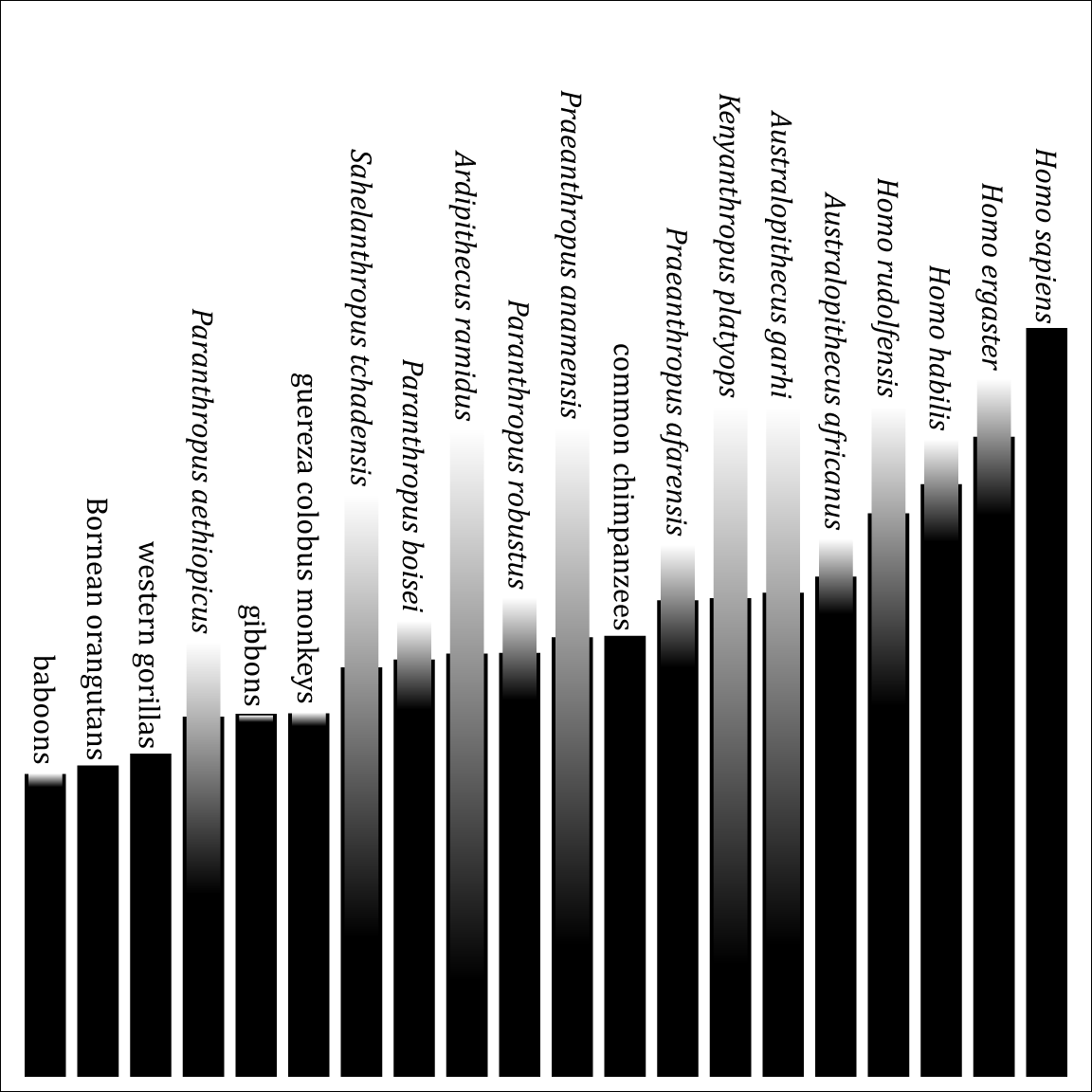
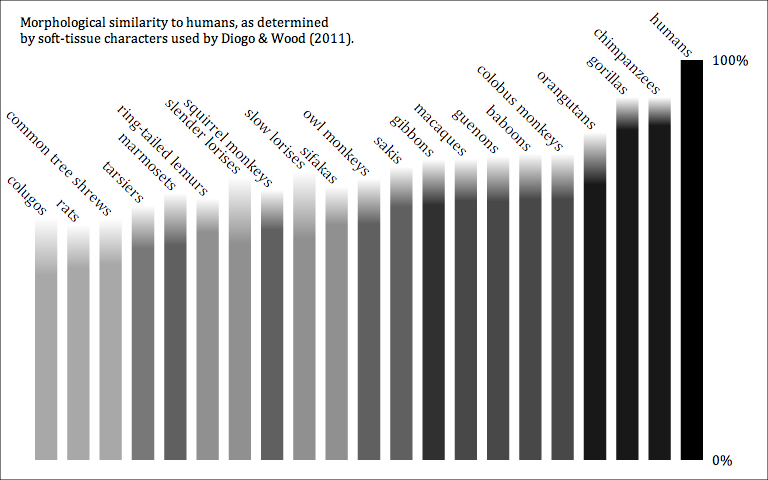
A genus is not an empirical entity. It's a bookkeeping convention, up to the personal whims of the taxonomist. And of course this leads to a huge mess.
Depending on the taxonomist (and, for early forms, the phylogeny), the human total group includes anywhere from one genus (
Homo) to ten (
Ardipithecus,
Australopithecus,
Homo,
Kenyanthropus,
Orrorin,
Paranthropus,
Paraustralopithecus,
Praeanthropus,
Sahelanthropus, and
Zinjanthropus — not even mentioning obsolete ones like
Telanthropus and
Pithecanthropus). We could try to clean up this mess by creating phylogenetic definitions and reinterpreting the genera as clades, except that all of the type species are thought by at least some researchers to be ancestral forms (with the exceptions of
Homo sapiens, Paranthropus robustus, and
Zinjanthropus boisei). For example, if
Australopithecus is a clade that includes
Australopithecus africanus, then it might also include
Homo, and genera are not allowed to overlap under the
ICZN.
If we use all of the above genera, then the taxonomy looks like this:
- Ardipithecus
- Ardipithecus anamensis (Not in Praeanthropus, despite sometimes being synonymized with afarensis! Although it should be noted that ramidus is much better known now than in 2004, so this may have changed.)
- Ardipithecus ramidus
- Australopithecus
- Australopithecus africanus
- Homo
- Homo ergaster
- Homo habilis (although it is closer to garhi than to sapiens!)
- Homo sapiens
- Kenyanthropus
- Kenyanthropus garhi (!)
- Kenyanthropus platyops
- Kenyanthropus rudolfensis (although it is closer to ergaster and habilis than to platyops, it is still closer to platyops than to sapiens)
- Orrorin
- Orrorin tugenensis (not included in the study, but this is nomenclaturally where it would go unless found to be a synonym)
- Paranthropus
- Paraustralopithecus
- Paraustralopithecus aethiopicus
- Praeanthropus
- Sahelanthropus
- Sahelanthropus tchadensis
- Zinjanthropus
Not included: species that are not types and were not included in the study, like Ardipithecus kadabba (scrappy craniodental remains), Australopithecus sediba (hadn't been discovered in 2004), Homo heidelbergensis (pretty close to sapiens anyway), etc.
It must be said that Zinjanthropus and Paraustralopithecus are not that commonly used. If we remove Paraustralopithecus, then aethiopicus predictably falls into Zinjanthropus. If we remove Zinjanthropus as well, then both aethiopicus and boisei predictably fall into Paranthropus.
Praeanthropus is also not that widely used. If we remove that as well, we get:
- Ardipithecus
- Ardipithecus anamensis
- Ardipithecus ramidus
- Australopithecus
- Australopithecus afarensis
- Australopithecus africanus
- Homo
- Homo ergaster
- Homo habilis
- Homo sapiens
- Kenyanthropus
- Kenyanthropus garhi
- Kenyanthropus platyops
- Kenyanthropus rudolfensis
- Orrorin
- Paranthropus
- Paranthropus aethiopicus
- Paranthropus boisei
- Paranthropus robustus
- Sahelanthropus
- Sahelanthropus tchadensis
Kenyanthropus is a rather controversial genus. If we remove it, we get:
- Ardipithecus
- Ardipithecus anamensis
- Ardipithecus garhi
- Ardipithecus platyops
- Ardipithecus ramidus
- Australopithecus
- Australopithecus afarensis
- Australopithecus africanus
- Australopithecus rudolfensis (still refuses to go with sapiens!)
- Homo
- Homo ergaster
- Homo habilis
- Homo sapiens
- Orrorin
- Paranthropus
- Paranthropus aethiopicus
- Paranthropus boisei
- Paranthropus robustus
- Sahelanthropus
- Sahelanthropus tchadensis
If we also remove Sahelanthropus, then tchadensis goes easily into Ardipithecus. I assume tugenensis would as well, if we removed Orrorin, but that wasn't included in the study since the craniodental material is so scant. If we remove Paranthropus, its species go very, very reluctantly into Australopithecus (by which I mean despite not being that close to africanus):
 |
Sahelanthropus tchadensis,
from PhyloPic |
- Ardipithecus
- Ardipithecus anamensis
- Ardipithecus garhi
- Ardipithecus platyops
- Ardipithecus ramidus
- Ardipithecus tchadensis
- Ardipithecus tugenensis?
- Australopithecus
- Australopithecus aethiopicus
- Australopithecus afarensis
- Australopithecus africanus
- Australopithecus boisei
- Australopithecus robustus
- Australopithecus rudolfensis
- Homo
- Homo ergaster
- Homo habilis
- Homo sapiens
Ardipithecus ramidus was originally named as Australopithecus ramidus. If we remove Ardipithecus, its species predictably end up in Australopithecus:
- Australopithecus
- Australopithecus anamensis
- Australopithecus aethiopicus
- Australopithecus afarensis
- Australopithecus africanus
- Australopithecus boisei
- Australopithecus garhi
- Australopithecus platyops
- Australopithecus ramidus
- Australopithecus robustus
- Australopithecus rudolfensis
- Australopithecus tchadensis
- Australopithecus tugenensis?
- Homo
- Homo ergaster
- Homo habilis
- Homo sapiens
Now for the final cut. What happens when we remove Australopithecus?
- Gorilla
- Gorilla aethiopicus (!!! although gorilla only beats sapiens by a hair)
- Gorilla beringei
- Gorilla garhi (!!!)
- Gorilla gorilla
- Gorilla tchadensis (admittedly, this was suggested by Senut and Pickford)
- Homo
- Homo africanus
- Homo boisei
- Homo ergaster
- Homo habilis
- Homo platyops
- Homo robustus
- Homo rudolfensis
- Pan
- Pan afarensis (!!)
- Pan anamensis (!!)
- Pan paniscus
- Pan ramidus (!!)
- Pan troglodytes
- incertae sedis
Yes, some of the stem-humans get pulled in with chimpanzees or gorillas! And there seems to be little rhyme or reason as to which go where. The splitting of the "robust australopithecines" is most bizarre (although, as noted, it only takes a tiny nudge to put aethiopicus in Homo with the others).
In summary:
- This is just one matrix, only focusing on one area of the anatomy.
- There are huge amounts of uncertainty with some of these taxa.
- Even without the uncertainty, there is no objective way to measure morphological distance.
- Even if there were, it might not be a good idea to use it to determine generic boundaries.
- Generic boundaries are stupid, anyway.