Despite the title of this blog, I've rarely if ever actually discussed simian brains. (The title refers to my own brain, or more generally the brain of any human that's ever thought about their place in the universe.) Reader Allison Gamble has kindly volunteered to rectify this situation with A Three-Pound Monkey Brain's first-ever guest post. Enjoy!
Similarities Between Macaque and Human Brains
by Allison Gamble
 |
Rhesus macaque
mother and child
by David Lewis |
Evolutionary scientists believe that humans and Old World monkeys, such macaques, evolved from a common ancestor. At some point in the past our paths diverged. As different as we may look on the outside, humans and macaques share many common behavioral traits, all of which have a basis hidden deep within our brains.
Rhesus macaques and other Old World monkeys are often used in scientific and medical experimentation. They make ideal research subjects because in many ways, human brains and Old World monkey brains are very similar. The more we learn about the similarities and differences between human and Old World monkey brains, the better the picture we can get of the two groups’ evolutionary paths since diverging from that common ancestor about 30 million years ago. What's more, these Rhesus macaques’ brains may hold keys to understanding the neurological and cognitive bases of prejudice, racial discrimination, and associated violent behaviors that could be massive boons to forensic psychology and other disciplines we humans use to better understand ourselves.
Size Matters
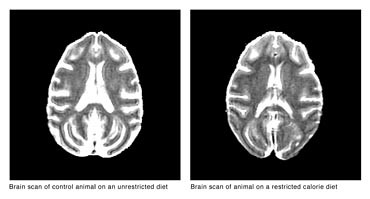 |
Scans of Rhesus macaque brains
by Ellery Chen |
What is the primary difference between Old World monkey brains and human brains? Size and shape are the most obvious. The average adult human brain weighs between 1,300 and 1,400 grams, and even an average newborn human baby's brain is about 350 to 400 grams. An average Rhesus macaque's brain is approximately 90 to 97 grams. Humans have the largest brain-to-body ratio of any primate.
A Single Gene Makes a Huge Difference
 |
Skeleton of 'Able',
a Rhesus macaque sent into space.
From the Otis Historical Archives,
National Museum of
Health & Medicine. |
According to a recent study conducted at Emory University, Rhesus macaques demonstrated a trait previously thought to belong solely to humans: recalling past images and applying them to a current situation. Why is this important? The ability to learn from past actions is the basis for imagination and planning for the future, the key to complex societies and survivability of the species. A Yale University study found the primary difference between Old World monkey brains and human brains may be the activation of a single gene, NDE1, which controls the growth of the cerebral cortex.
The cerebral cortex covers both hemispheres of the brain. While parts of it control basic physiological functions like sensory input and limb or eye movement, other areas are responsible for advanced functions like memory, abstract thinking, language, creativity, emotion, judgment and attention. While initially it may seem like these are solely human abilities, studies and research have shown that this idea is false.
Macaques Have Self-Doubt
As an example, one study found that macaques have the capacity to doubt themselves in a manner similar to humans. The professors conducting the study, Michael Beran from Georgia State University and John David Smith from State University of New York, trained macaques to play a computer game that asked the macaques questions. The macaques were given a treat when they answered the questions correctly. Over time, the macaques learned to avoid the difficult questions. When they were unsure of their answer, they could pass on the question. New World monkeys were also tested, but did not show the same awareness of their own thought. Since New World monkeys are not as similar to us as Old World monkeys this was to be expected. The significance of the study, according to Dr. Smith, is that self-awareness is a crucial part of human development, and the presence of it in Old World monkeys could help provide a sense of how it evolved.
Macaques Have Mirror Neurons
Both humans and macaques have what are known as 'mirror neurons.' These brain cells fire when we observe someone else performing the same action as us. What is the significance of mirror neurons? They're crucial to the capacity for behaviors such as empathy, emotional contagion (for example, when others laugh and you can't help but start laughing as well), and even developmental disorders like autism.
Instincts in Humans
 |
Rhesus macaque
by Vincent van Dam |
Some scientists don't like to use the word 'instincts' in association with humans. They prefer terms like 'natural predisposition.' While this may simply be a semantic difference, it is the cause of much controversy and debate in scientific circles.
Do humans have an instinct for violence? If so, where did that instinct come from? The answer isn't simple, as the nature vs. nurture debate has been raging for ages. One study indicated that Rhesus macaques recognize other Rhesus macaques as either belonging to the group or as outsiders. They also appeared to associate the outsiders with things that were frightening or dangerous, leading the researcher to conclude that the macaques had instincts for prejudice and even racism. By extension, humans may also have a disposition towards racism and prejudice, two behaviors that have certainly inspired a fair share of violence over the years.
By continuing to study our Old World cousins, we can discover much about our own behavior, including how we learn, how we view others and, how we view ourselves. However, despite the striking similarities between human and Old World monkey cognition and behavior, a crucial difference in humans remains that once we are aware of how we behave, we can self-correct. In this way we separate ourselves from the monkeys, and perhaps it is this ability that more than anything else defines our humanity.