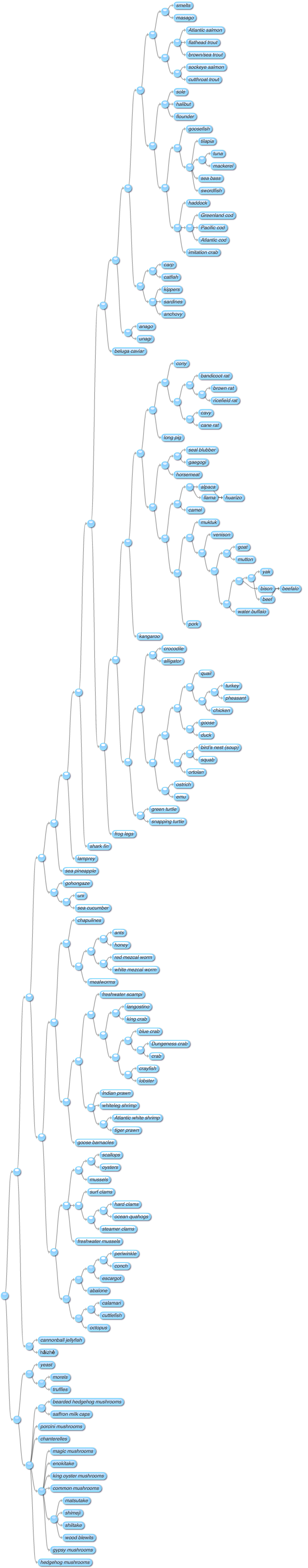I just ran Radish on Zea mays (maize, a.k.a. corn). Look what the combined taxonomies from uBio look like:
What a mess! Keep in mind that the multiplicity of paths is not due to differing phylogenies (they all seem to agree on that), but to differing nomenclature. Even if uBio were to add some of the more obvious synonymies (e.g., Embryophyta and "Embryophytes"), it'd still be pretty wild.
Eventually I plan to have Radish work with automatically-generated taxonomies, made by placing PhyloCode names (from RegNum) onto TreeBase phylogenies using Names on Nodes algorithms, but until then I guess this is the best option.
And Zea mays is just a particularly egregious example. In contrast, here's a nice, neat "radish" for Scarabaeus sacer:
Apart from the one errant use of Animalia, pretty nice!
17 August 2010
Radish: Pulling Up Taxonomic Hierarchies From Leaf to Root
I've just released a small open-source Java library called Radish. It contains tools for interfacing with uBio (and hopefully other services in the future), with the purpose of looking up a taxon's hierarchy. It does this by synthesizing multiple taxonomies into a single graph. Here, for example, is the graph produced by looking up Homo sapiens Linnaeus 1758 (click the image to enlarge):
This is a synthesis of several taxonomies. Some have more ranks than others, but they all converge on a single chain, with the exception of the taxa above the "phylum level", where at least two of them use different schemes. (Although it may be tempting to synonymize Biota and "Cellular life", or Animalia and Metazoa, these are not objective synonymies, and arguably the former is a bad idea anyway.) Ideally the library would be able to figure out that "Cellular life" includes Biota (if the latter is a crown clade), and that Animalia either includes or is a synonym of Metazoa, but it can only work with the data it's given.
Why the name "Radish"? From the project's wiki:
This is a synthesis of several taxonomies. Some have more ranks than others, but they all converge on a single chain, with the exception of the taxa above the "phylum level", where at least two of them use different schemes. (Although it may be tempting to synonymize Biota and "Cellular life", or Animalia and Metazoa, these are not objective synonymies, and arguably the former is a bad idea anyway.) Ideally the library would be able to figure out that "Cellular life" includes Biota (if the latter is a crown clade), and that Animalia either includes or is a synonym of Metazoa, but it can only work with the data it's given.
Why the name "Radish"? From the project's wiki:
The English word "radish" is derived from Latin "radicem" (nominative: "radix"), meaning "root". (This is also where the word "radical" comes from.) The idea of the Radish library is that by "grabbing a leaf" (i.e., selecting a smaller taxon), you can "pull up the root" (i.e., extract that taxon's hierarchy of supertaxa, up to the root of all life).Radish is part of a larger project I am working on ... I should have more to say about that in the near future....
02 August 2010
Phun Phylogenies
Pete Buchholz recently started compiling a phylogeny of edible plants, based on the APG III system. I ran it through Names on Nodes and produced a diagram:
(Unfortunately, this version strips out the clade labels—I'll try and rectify that at some point.)
When I saw this, I though, what a fantastic way to learn plant phylogeny! It's something I don't know much about (apart from basics, like the differerence between gymnosperms and angiosperms), and so I found it fascinating to see the ways the foods I eat are related to each other.
It's such a good idea, I couldn't resist doing another version for edible animals and fungi:
And this reminded me of another project I'd been meaning to start for a while, so I finally took a stab at it. A phylogeny of cartoon animals!
(That's right, there's a stuffed tiger clade.)
(Unfortunately, this version strips out the clade labels—I'll try and rectify that at some point.)
When I saw this, I though, what a fantastic way to learn plant phylogeny! It's something I don't know much about (apart from basics, like the differerence between gymnosperms and angiosperms), and so I found it fascinating to see the ways the foods I eat are related to each other.
It's such a good idea, I couldn't resist doing another version for edible animals and fungi:
And this reminded me of another project I'd been meaning to start for a while, so I finally took a stab at it. A phylogeny of cartoon animals!
(That's right, there's a stuffed tiger clade.)
Labels:
animation,
cartoons,
charts,
clade,
diagrams,
gastronomics,
Names on Nodes,
phylogeny
Subscribe to:
Posts (Atom)