A while back I started the March of Man project. The goal is to create a massive illustration of human evolution using hundreds of figures. My first attempt involved drawing them by hand on a big piece of paper. I think that lasted about a week.
My second attempt involved a rich web application, where people could submit image files according to certain specifications, and these would be incorporated into a big collage. This got a bit further, but went stagnant for a few reasons: 1) the application is a bit buggy and unwieldy (it was my first big Flex project), 2) drawing each figure still takes a while, and 3) I overestimated the amount of public interest there would be in contributing to this project. (Thanks to Steve O'Connor, though, for being the only other contributor!)
It's been a couple of years since I launched that site, and now I've started on a new method. Will it go anywhere? We'll see. It does solve some problems of the previous approaches.
This time, I'm doing it in 3D. The plan: 1) create a poseable model of an Ardipithecus ramidus female; 2) create morph targets for various other species, ages, and for males; 3) use this to create images for significant localities; 4) combine these into one vast mural.
So far I'm almost done with step #1. Here she is:
(Also posted on my sketch blog, Dragabok.)
Still have to tweak the hair a bit and make her poseable. After that, I've got my work cut out for me....
01 December 2009
The Mangani Clade
As I've mentioned earlier, the formal nomenclature for apes (including humans) is a huge mess. One person's "hominin" is another's "hominid" and my "hominine" might be your "homininan". And in this situation I honestly think the vernacular terms (adjusted in some cases to include humans) serve us better:
There is one possible vernacular term I've seen for this clade, and it comes from an unexpected source.

Tarzan is one of the most popular and enduring fictional creations of the 20th century. Everyone knows he was raised by apes—but what kind of apes? In various films they are depicted as gorilla-like (e.g., Disney's Tarzan) or chimpanzee-like (e.g., Greystoke: The Legend of Tarzan, Lord of the Apes). What were they in the novels?
The author, Edgar Rice Burroughs, never attempted to identify them scientifically. In the novels they have a primitive, guttural language, and they call themselves mangani. They are not gorillas, because they have another word for those (bolgani). Chimpanzees are never mentioned in the novels.
Interestingly, the mangani also use the word mangani for humans. For example, black people are gomangani and white people are tarmangani. So, in their own self-taxonomy, there is a group that includes themselves and humans, but excludes gorillas. Sound familiar?
The mangani can't be chimpanzees, because they are much larger (although less massive than gorillas). Like chimpanzees, they are fairly arboreal, but, unlike them, they do not use tools (apart from logs as drums, perhaps), do speak a language, and do not hunt cooperatively. Still, considering that chimpanzee behavior in the wild was virtually unknown when Burroughs was writing, and that chimpanzees were not thought to be closer to humans than to gorillas, perhaps he can be given some leeway here, since his prescience is otherwise impressive. (Especially given the recent discovery of a possible population of giant chimpanzees in the Bili Forest of the Congo.)
I'd love to learn of a good vernacular term for the human-chimpanzee crown clade, but until such a time as I do (or the formal nomenclature becomes actually useable), I like the idea of referring to humans, common chimpanzees, bonobos, Ardipithecus, "Lucy", Australopithecus, Floresian "hobbits", Homo erectus, Neandertals, etc. as "mangani".
- apes: gibbons, great apes
- gibbons (or lesser apes): Hoolock, Hylobates, Nomascus, Symphalangus
- great apes: African apes, Pongo
- African apes: Gorilla, Homo, Pan
There is one possible vernacular term I've seen for this clade, and it comes from an unexpected source.

Tarzan is one of the most popular and enduring fictional creations of the 20th century. Everyone knows he was raised by apes—but what kind of apes? In various films they are depicted as gorilla-like (e.g., Disney's Tarzan) or chimpanzee-like (e.g., Greystoke: The Legend of Tarzan, Lord of the Apes). What were they in the novels?
The author, Edgar Rice Burroughs, never attempted to identify them scientifically. In the novels they have a primitive, guttural language, and they call themselves mangani. They are not gorillas, because they have another word for those (bolgani). Chimpanzees are never mentioned in the novels.
Interestingly, the mangani also use the word mangani for humans. For example, black people are gomangani and white people are tarmangani. So, in their own self-taxonomy, there is a group that includes themselves and humans, but excludes gorillas. Sound familiar?
The mangani can't be chimpanzees, because they are much larger (although less massive than gorillas). Like chimpanzees, they are fairly arboreal, but, unlike them, they do not use tools (apart from logs as drums, perhaps), do speak a language, and do not hunt cooperatively. Still, considering that chimpanzee behavior in the wild was virtually unknown when Burroughs was writing, and that chimpanzees were not thought to be closer to humans than to gorillas, perhaps he can be given some leeway here, since his prescience is otherwise impressive. (Especially given the recent discovery of a possible population of giant chimpanzees in the Bili Forest of the Congo.)
I'd love to learn of a good vernacular term for the human-chimpanzee crown clade, but until such a time as I do (or the formal nomenclature becomes actually useable), I like the idea of referring to humans, common chimpanzees, bonobos, Ardipithecus, "Lucy", Australopithecus, Floresian "hobbits", Homo erectus, Neandertals, etc. as "mangani".
Labels:
anthropology,
nomenclature,
primatology,
taxonomy
20 November 2009
More Human Evolution Diagrams
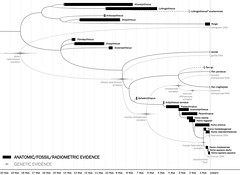
I'm nearing the end of work on a review of the case for human evolution. I've uploaded some of the diagrams to my Flickr account before. Some of these were just updated and some new ones were added: see The Case for Human Evolution (Flickr set).
This is probably my favorite of the lot, showing the congruence between morphological/paleontological data (including radiometric dates) and genetic data (including molecular clocks):
Can you spot the single discrepancy? (No fair reading the caption on Flickr first.)
UPDATE: Revised some of the images, temporarily removed one.
Labels:
charts,
diagrams,
evolution,
illustration,
paleontology,
phylogeny
What I Do For a Living, Part 312: Dr. Facilier's Parlour
I recently completed a new application. This one is a collection of three games associated with Disney's upcoming animated film, The Princess and the Frog. This one requires a Facebook account: Dr. Facilier's Parlour. (Dr. Facilier is the villain of the movie.)
This is my first completed project using certain technologies:
Also see the Shadow Shakedown game therein for an example of how
I have to say I was very pleased with all of these new technologies. In particular, the Spark component set is a huge improvement over the previous Halo components. Skinning components is so easy now it's hard to imagine that it was ever difficult. Adobe has acted on the "Favor composition to inheritance" maxim, and it has paid off. Also, props to Code Igniter, with its flexible tools and strong emphasis on the MVC pattern, for making PHP development (something I had all but sworn off) actually kind of fun.
Enjoy!
This is my first completed project using certain technologies:
- Facebook API
- Spark components (Flex 4 beta)
- Code Igniter PHP framework
Also see the Shadow Shakedown game therein for an example of how
DisplacementMapFilter can be useful. (I also got to do a tiny bit of character animation there.)I have to say I was very pleased with all of these new technologies. In particular, the Spark component set is a huge improvement over the previous Halo components. Skinning components is so easy now it's hard to imagine that it was ever difficult. Adobe has acted on the "Favor composition to inheritance" maxim, and it has paid off. Also, props to Code Igniter, with its flexible tools and strong emphasis on the MVC pattern, for making PHP development (something I had all but sworn off) actually kind of fun.
Enjoy!
Labels:
ActionScript,
animation,
Facebook,
film,
Flash,
Flex,
PHP,
RIAs,
social media,
web applications,
work
07 September 2009
Glimpses of Stuff I've Been Working On
Two long-term projects of mine should see the light of day soon. Here are some previews (click to see full size):
Labels:
definition,
evolution,
MathML,
Names on Nodes,
phylogeny
24 August 2009
Online NEXUS File Viewer
It's been a month since my last post, but I have a very good reason for the hiatus. Namely, I was busy getting married to this woman (at the Los Angeles County Museum of Natural History) and going on our honeymoon (in Sydney, Australia).
Now that I'm back in California, time to get back to work on Names on Nodes! I've just put together a small demo of two key parts of its functionality: the reading of NEXUS files and the displaying of phylogenetic networks. Click here to see the NEXUS Viewer demo. This application opens NEXUS files and displays the trees in them as a combined phylogenetic network.
Things you need to know:
- You must have a NEXUS file stored locally on your computer to use this.
- That file should have a TREES section. (If not, the viewer should just display a list of operational taxonomic units.)
- This could get messy for NEXUS files with lots of trees. (Although it's kind of neat-looking.)
- You can move the nodes around by clicking on them, or click anywhere else to move the entire diagram.
- I would dearly love to know if, for some reason, it does not work for a given file.
Enjoy!
Labels:
ActionScript,
cladistic analysis,
evolution,
Flash,
Flex,
Names on Nodes,
personal,
phylogeny,
RIAs,
tools,
web applications
23 July 2009
Two "Names on Nodes"-Related Launches
I'm still a clear way away from launching the beta application, but I've just made a couple of launches related to my long-time work-in-progress, Names on Nodes.
First up, and probably of more interest to most people, I've begun the documentation for the MathML definitions used by Names on Nodes. The document includes general reviews of relevant mathematical and biological concepts, a quick review of MathML and the technologies it's based on, some comments on correlating mathematical and biological concepts, and definitions for all entities (including operations) used by Names on Nodes. Note that this covers a lot of the same ground as in my 2007 paper, with a few minor changes in the symbols and terminology (e.g., I now call the ancestor of a clade a "cladogen" rather than a "cladogenetic set").
Secondly, I've made the project open-source, by moving it to Google Code. If you are a developer interested in checking this out, go here. It's incomplete, so I don't know if anyone will have any real interest in looking at it yet. (Honestly, I mostly posted so that, on the off chance that I unexpectedly kick the bucket, my magnum opus won't be lost forever.)
This information is also on the new Names on Nodes home page.
Labels:
ActionScript,
databases,
definition,
Flash,
Flex,
Java,
mathematics,
MathML,
MXML,
Names on Nodes,
nomenclature,
open source,
phylogeny,
RIAs,
systematics,
taxonomy,
web applications,
XML
22 July 2009
"The Case for Human Evolution" - Illustrations
I have been working on an essay entitled The Case for Human Evolution for a while. I've just posted some illustrations I've been working on:
Enjoy!
30 June 2009
New Useless Utility: Text Tree Maker
I finally got around to launching something at namesonnodes.org. No, it's not Names on Nodes itself, unfortunately. The project is taking a huge amount of time. But I thought I'd post something, so here's a little Flex application I made (using the new Flash Builder 4 Beta!) using a smidgen of the technology behind Names on Nodes.
Have you ever been discussing phylogeny online and wished there was an easy way to make a readable cladogram? (95% of readers leave.) Those of you who are left, check this out: Text Tree Maker. Just type in a Newick tree string, and voilá! Okay, so typing in a Newick tree string is not that easy in the first place, but it is easier.
Well, I'll be using it, anyway. Check this one out!
NOTE: Right-click on the application and select "View Source" if you want to see some of the code behind it.
Have you ever been discussing phylogeny online and wished there was an easy way to make a readable cladogram? (95% of readers leave.) Those of you who are left, check this out: Text Tree Maker. Just type in a Newick tree string, and voilá! Okay, so typing in a Newick tree string is not that easy in the first place, but it is easier.
Well, I'll be using it, anyway. Check this one out!
Ardipithecus
|--ALA-VP 2/10
`--+--ARA-VP 6/1
|--KNM-T1 13150
`--Praeanthropus
|--KNM-KP 29281
`--+--AL 288-1
|--KNM-WT 40000
|--KT 12/H1
|--LH 4
`--+--BOU-VP 12/130
|--Australopithecus
| |--Taung 1
| |--Australopithecus (Paranthropus)
| | |--SK 6
| | `--TM1517
| `--Australopithecus (Zinjanthropus)
| |--KNM-WT 17000
| |--OH 5
| `--Omo 18
`--Homo
|--KNM-ER 1470
|--KNM-ER 1813
|--OH 7
|--OH 9
`--Homo (Homo)
|--D 2600
|--KNM-ER 992
|--LB 1
`--+--Ceprano 1
|--Trinil 2
`--Homo (sapiens)
|--ATD 6-5
`--+--Mauer 1
`--+--Neandertal 1
`--+--Florisbad 1
|--Kabwe 1
`--Uppsala domkyrka: Carolus Linnaeus
NOTE: Right-click on the application and select "View Source" if you want to see some of the code behind it.
Labels:
anthropology,
Flash,
Flex,
Names on Nodes,
RIAs,
web applications
24 June 2009
Human-Chimpanzee Systematics
I've been working on a couple of projects to do with stem-humans. Naturally, these efforts necessitate creating a working phylogeny. I thought I'd post what I more or less have so far. I haven't done any rigorous work here; I'm just trying to piece things together from various publications.
This is a phylogeny of all known species within Clade(Homo sapiens Linnaeus 1758 ← Troglodytes gorilla Savage vide Savage & Wyman 1847), including some unnamed, fragmentary species that can only be differentiated from other species by location and/or time. (Note: Sahelanthropus tchadensis Brunet & al. 2002 is excluded because it doesn't seem to be clear that it does fall within this clade.) I've included links for all citations with permanent identifiers, when available, or popups with fuller information, when not. The phylogeny is interspersed with a rank-based taxonomy. (Unfortunately, there are no published phylogeny-based names to apply here.) Outlined circles indicate that the species may be ancestral to what are shown as sister groups. Species names are listed with their original prenomina (genera), regardless of current placement. I've added a note when the listed species is the type of its prenominal genus or another genus.
This is a phylogeny of all known species within Clade(Homo sapiens Linnaeus 1758 ← Troglodytes gorilla Savage vide Savage & Wyman 1847), including some unnamed, fragmentary species that can only be differentiated from other species by location and/or time. (Note: Sahelanthropus tchadensis Brunet & al. 2002 is excluded because it doesn't seem to be clear that it does fall within this clade.) I've included links for all citations with permanent identifiers, when available, or popups with fuller information, when not. The phylogeny is interspersed with a rank-based taxonomy. (Unfortunately, there are no published phylogeny-based names to apply here.) Outlined circles indicate that the species may be ancestral to what are shown as sister groups. Species names are listed with their original prenomina (genera), regardless of current placement. I've added a note when the listed species is the type of its prenominal genus or another genus.
- ?Orrorin tugenensis Senut & al. 2001 [typus]
- ≅ Tribus Hominini Gray 1825
- Ardipithecus kadabba Haile-Selassie 2001
- ≅ Genus Pan Oken 1816
- Pan sp. innom. McBrearty & Jablonski 2005
- Pan paniscus Schwarz 1929
- Simia troglodytes Blumenbach 1776 [typus Pan]
- ≅ Subtribus Hominina Gray 1825
- Australopithecus ramidus White & al. 1994 [typus Ardipithecus]
- Australopithecus anamensis Leakey & al. 1995 (?= praegens)
- Homo praegens Ferguson 1989
- Australopithecus afarensis Johanson & al. 1978 [typus Praeanthropus]
- Australopithecus bahrelghazali Brunet & al. 1995 (?= afarensis)
- Australopithecus garhi Asfaw & al. 1999
- Kenyanthropus platyops Leakey & al. 2001 [typus] (?= afarensis)
- ≅ Genus Australopithecus Dart 1925
- Australopithecus aethiopicus Olson 1985
- Australopithecus africanus Dart 1925 [typus]
- Zinjanthropus boisei Leakey 1959 [typus]
- Australopithecus robustus Broom 1938 [typus Paranthropus]
- ≅ Genus Homo Linnaeus 1758
- Homo sp. innom. Kimbel & al. 1996
- Homo habilis Leakey & al. 1965
- Homo rudolfensis Alexeev 1986
- ≅ Subgenus Homo (Homo) Linnaeus 1758
- Anthropopithecus erectus Dubois 1892 [typus]
- Homo ergaster Groves & Mazák 1975
- ?Homo floresiensis Brown & al. 2004
- Homo georgicus Vekua & al. 2002
- ≅ Superspecies Homo (sapiens) Linnaeus 1758
- Homo antecessor Bermúdez de Castro & al. 1997
- Homo cepranensis Mallegni & al. 2003 (?= antecessor)
- Homo heidelbergensis Schoetensack 1908
- Homo neanderthalensis King 1864
- Homo rhodesiensis Woodward 1921 (?= heidelbergensis)
- Homo sapiens Linnaeus 1758 [typus]
Labels:
anthropology,
evolution,
mammalia,
nomenclature,
paleontology,
phylogeny,
primatology,
systematics,
taxonomy,
Vertebrata,
zoology
29 May 2009
One Name, One Taxon
One of the primary goals of a nomenclatural code should be to make sure that names refer to one taxon and one taxon only. This principle is mentioned in the ICZN's preamble (emphasis added):
The objects of the Code are to promote stability and universality in the scientific names of animals and to ensure that the name of each taxon is unique and distinct. All its provisions and recommendations are subservient to those ends and none restricts the freedom of taxonomic thought or actions.This point is reiterated in Article 52:
52.1. Statement of the Principle of Homonymy. When two or more taxa are distinguished from each other they must not be denoted by the same name.Yet there are numerous cases in zoological nomenclature where this rule is flagrantly ignored. A few:
- "Echinoidea" is the name of a superfamily containing Echinus (a sea urchin genus), but also the name of a class containing that superfamily.
- "Ophiuroidea" is the name of a superfamily containing Ophiura (a brittle star genus), but also the name of a class containing that superfamily.
- "Chelonia" is a genus of turtle, but also used as the name of the order containing all turtles.
- "Pterodactyloidea" is the name of taxon given various ranks (usually suborder) including most short-tailed pterosaurs, but also the name of a superfamily within that taxon.
This is getting to be an actual problem for me, because parts of Names on Nodes rely on the principle that a name only has one meaning under a given authority. When I create a database entry for urn:isbn:0853010064::Pterodactyloidea, is it for a suborder or a superfamily? ICZN rules actually dictate that the superfamily has precedence, since Family Pterodactylidae Meyer 1830 has precedence over Suborder Pterodactyloidea Plieninger 1901. (The ICZN considers the naming of any taxon whose rank is in the family group as implicitly naming taxa for all ranks of the family group; thus, naming Family Pterodactylidae implicitly names Superfamily Pterodactyloidea, Subfamily Pterodactylinae, Tribe Pterodactylini, and Subtribe Pterodactylina.) People who use "Pterodactyloidea" for a suborder, beware! You are violating the rules of the ICZN! (WhooOOOOoo!!)
The situation with "Echinoidea" is even worse. As near as I can tell, Family Echinidae was named by Gray in 1825 (thus implicitly naming Superfamily Echinoidea), but Class Echinoidea was already named by Leske in 1778. And the ICZN mandates that the superfamily including Echinus must be named "Echinoidea" if the family is named "Echinidae". I'm not sure how this is supposed to play out ... does Echinus simply not get a name for its superfamily? Those poor wee urchins....
(And people wonder why I support an alternative nomenclatural code without mandated suffixes for ranks!)
In the case of Chelonia, people are generally using another name ("Testudines") for the order nowadays, but in other cases I've got a real problem, especially if I hope to automatically pull a lot of this data from other databases.
Labels:
databases,
Names on Nodes,
nomenclature,
taxonomy
27 May 2009
PLoS ONE exPLoSiONE
Others have said it before me, but I think PLoS ONE and online journals like it represent the future of scientific publishing. Quick turnaround, open access, unlimited space (not just for text, but also images, data files, etc.)—I just don't see how the older forms of journal can possibly persist for long.
Perhaps the most useful feature, though, is sadly underutilized. Imagine this—you're reading a paper and you come across an error, or a questionable inference, or an unclear point. With printed journals, you have the following options:
- Write to the editor and/or primary author and hope they have a moment to respond.
- Complain to whomever will listen.
- Write a frustrated note in the margin and move on.
- Fume silently.
But with PLoS ONE, you can accomplish #1–3 all at once. (And #4, if you want.) Simply log in, highlight the text you want to comment on (or click on "Leave a general comment"), and you can leave a publicly-visible note that anyone, authors and editors included, can respond to.
People have been slow to take advantage of this wonderful system, but it's starting to take off, I think. As many are aware, there's been a big media hubbub about a new fossil primate (Darwinius masillae) that may or may not be a stem-haplorhine (i.e., part of the group that gave rise to tarsiers and monkeys, including apes, including humans). I've seen a lot of discussion of it in various venues, and some of that is finally starting to spill over into the paper's comments section. (You may note a couple of comments left by myself.)
One notable outcome of the discussion on Darwinius is the rectification of an incompatibility between PLoS ONE's publication methods and the current requirements of the ICZN. This meant that the new scientific names published in the journal (e.g., "Darwinius", "Darwinius masillae") were nomenclaturally unavailable. Happily, this was quickly resolved, and the remedy was also carried out for the names introduced in some earlier papers.
Other discussions, on such topics as the scoring of characters as "Derived" or "Primitive", are still ongoing.
PLoS ONE also allows readers to rate the articles and leave reviews. (As of this writing, there is one review by Andy Farke, and I think he makes some excellent points.)
Science open to everyone! Go ahead—get involved.
Labels:
nomenclature,
paleontology,
primatology,
publication,
Vertebrata,
zoology
05 May 2009
'Nother Toolshop Animatic: The Head Map
I created a sort of "mashup" of the previous two and added a temporary music track. (The ultimate version will have something different.) Click on the thumbnail (might take a moment to load):

Still clunky, but I'm just fleshing out the ideas at this point. Enjoy!

Still clunky, but I'm just fleshing out the ideas at this point. Enjoy!
Labels:
animation,
anthropology,
evolution,
Flash,
mammalia,
March of Man,
primatology
04 May 2009
March of Man: The Toolshop
My somewhat ambitious web app, March of Man, has not been proving too successful. The idea behind the project is to illustrate human and chimpanzee evolution using hundreds of figures. The web app includes tools for submitting images and generating collages. But there are only a couple dozen images right now. At this rate, the project will be completed by the time I am an old man. Time for a new approach!
I'm going to leave the site up as is, but I am also going to be working on a CG animation. I've made a new area of the website called "The Toolshop" where I'll be posting progress. Here are the first two mockups, using vector animation (click on the image to see the animation):
I'm going to leave the site up as is, but I am also going to be working on a CG animation. I've made a new area of the website called "The Toolshop" where I'll be posting progress. Here are the first two mockups, using vector animation (click on the image to see the animation):
Labels:
animation,
anthropology,
evolution,
Flash,
mammalia,
March of Man,
primatology,
zoology
23 March 2009
Logging In Without a Password
I probably have hundreds of online accounts: email, discussion forums, social networking, online shopping, server hosting, issue reporting, etc. Trying to remember all the passwords is a pain. Often, when going to a site I haven't been to in a while, I just reset it, or have it sent by email, or have a new one sent by email, or however the site in question works.
I might want to try something different for Names on Nodes. As hinted at in earlier posts, users will be considered "authorities" in Names on Nodes, along with publications, bioinformatics files, specimen repositories, nomenclatural codes, etc. All authorities are associated with one or more unique URIs, such as website addresses, ISBN numbers, DOIs, LSIDs, etc. For users, the primary URI will be an email address, in the form <mailto:myname@somedomain.tld>.
Why have an account? Well, because then, as an authority, you get to "authorize" your own datasets and taxon identifiers (and, by proxy, taxon definitions). Datasets and taxon identifiers are "qualified" objects, meaning that they each refer to an authority, and they each have a "local name" unique under that authority. A qualified name is formed by joining the URI of the authority and the local name. So, for example, if you wanted to create a new phylogenetic hypothesis about mushrooms, it might have the qualified name <mailto:me@myemailprovider.com::dataset:basidiomycota+phylogeny>. If you wanted to provide your own definition for the name "Eumetazoa", it would be attached to a taxon identifier with the qualified name <mailto:me@myemailprovider.com::Eumetazoa>. And so on.
How do you log in without a password? I'm thinking of a system involving IP addresses, the numerical code that identifies your computer's connection. For most environments, these are relatively stable, although if you use, e.g., a DSL modem it may reset once in a while. Here are some potential use cases:
Preconditions.—User has never logged in. User's email is unregistered. User is 13 years of age or older.
Trigger.—User tries to do something that requires login.
Course of events.
Preconditions.—User's email is registered. User is not logged in, having logged out or having declined to stay logged in across sessions.
Trigger.—User tries to do something that requires login.
Course of events.
Preconditions.—User's email is registered. User indicated that they wanted to stay logged in across sessions the last time they logged in.
Trigger.—User visits website.
Course of events.
Preconditions.—User's email is registered. User has never logged in from their current IP address.
Trigger.—User tries to do something that requires login.
Course of events.
Preconditions.—User's email is registered, and user is logged in.
Trigger.—User decides to invalidate other IP addresses, perhaps fearing someone else may log in as them from another computer.
Course of events.
I'm not sure if this would be too convoluted in practice, but somehow I doubt it. If anything, it seems no worse then the usual type of system, except possibly for people who use laptops and are constantly on the move.
What do people think? Ideas, questions, concerns?
I might want to try something different for Names on Nodes. As hinted at in earlier posts, users will be considered "authorities" in Names on Nodes, along with publications, bioinformatics files, specimen repositories, nomenclatural codes, etc. All authorities are associated with one or more unique URIs, such as website addresses, ISBN numbers, DOIs, LSIDs, etc. For users, the primary URI will be an email address, in the form <mailto:myname@somedomain.tld>.
Why have an account? Well, because then, as an authority, you get to "authorize" your own datasets and taxon identifiers (and, by proxy, taxon definitions). Datasets and taxon identifiers are "qualified" objects, meaning that they each refer to an authority, and they each have a "local name" unique under that authority. A qualified name is formed by joining the URI of the authority and the local name. So, for example, if you wanted to create a new phylogenetic hypothesis about mushrooms, it might have the qualified name <mailto:me@myemailprovider.com::dataset:basidiomycota+phylogeny>. If you wanted to provide your own definition for the name "Eumetazoa", it would be attached to a taxon identifier with the qualified name <mailto:me@myemailprovider.com::Eumetazoa>. And so on.
How do you log in without a password? I'm thinking of a system involving IP addresses, the numerical code that identifies your computer's connection. For most environments, these are relatively stable, although if you use, e.g., a DSL modem it may reset once in a while. Here are some potential use cases:
Initial Login
Preconditions.—User has never logged in. User's email is unregistered. User is 13 years of age or older.
Trigger.—User tries to do something that requires login.
Course of events.
- User is prompted for their email address. They are also prompted on whether they want to stay logged in across sessions.
- User is prompted for their birthdate, full name, and family name.
- User gets a notice telling them that they have been sent a confirmation via email. The notice includes an input field for a "key".
- User checks their email, and sees an email message with a link. There is also a "key", a string of letters and numbers that they can copy and paste.
- User clicks on the link.
- Names on Nodes reopens, with the user logged in.
- User copies and pastes the "key" into the input field.
- User is now logged in.
Subsequent Login, Registered IP Address
Preconditions.—User's email is registered. User is not logged in, having logged out or having declined to stay logged in across sessions.
Trigger.—User tries to do something that requires login.
Course of events.
- User is prompted for their email address. They are also prompted on whether they want to stay logged in across sessions.
- User is now logged in.
Automatic Subsequent Login, Registered IP Address
Preconditions.—User's email is registered. User indicated that they wanted to stay logged in across sessions the last time they logged in.
Trigger.—User visits website.
Course of events.
- User is automatically logged in, and their name is shown in a "Welcome" message.
Subsequent Login, Unregistered IP Address
Preconditions.—User's email is registered. User has never logged in from their current IP address.
Trigger.—User tries to do something that requires login.
Course of events.
- User is prompted for their email address. They are also prompted on whether they want to stay logged in across sessions.
- User enters their email address.
- User gets a notice telling them that they have been sent a confirmation via email. The notice includes an input field for a "key".
- User checks their email, and sees an email message with a link. There is also a "key", a string of letters and numbers that they can copy and paste.
- User clicks on the link.
- Names on Nodes reopens, with the user logged in.
- User copies and pastes the "key" into the input field.
- User is now logged in.
Unregistering IP Addresses
Preconditions.—User's email is registered, and user is logged in.
Trigger.—User decides to invalidate other IP addresses, perhaps fearing someone else may log in as them from another computer.
Course of events.
- User selects the "Block Other Locations" option.
- User is prompted to confirm this request.
- User confirms the request.
- User receives notification that other locations have been unregistered.
I'm not sure if this would be too convoluted in practice, but somehow I doubt it. If anything, it seems no worse then the usual type of system, except possibly for people who use laptops and are constantly on the move.
What do people think? Ideas, questions, concerns?
17 March 2009
Refactoring "Names on Nodes" Entities, Part II
As I discussed previously, the Names on Nodes project had reached a point where the schema just wasn't working out. I went through a list of what was wrong with it: confusing nomenclature, various unnecessary classes, unnecessary references, and major practical problems with looking up contextual relations.
Another big problem was the home-brewed keyword search system I had going. Synchronizing the keyword lists was becoming problematic, and I realized there are already perfectly good (better, even) tools out there such as Hibernate Search. That's a chief rule of programming: don't reinvent something that people smarter than you, with more time on their hands, have already invented.
After a clear, honest look at the contextual relations, I came to a realization: they should be in the client, not the back end. No need to bog down the server with computing definition applications when it can be done in the client. That simplified things a great deal.
Another thing I didn't really need was categories. They were basically an ad hoc form of class inheritance, e.g., a species name is a nomen, a nomenclatural code is a publication, etc. For a little while I considered implementing this as a class hierarchy, as I had in earlier versions. But, really, this is irrelevant data—Names on Nodes doesn't really need to know what category an identifier falls in.
Finally, I had another problem in the way datasets and taxon identifiers (=signifiers) used qualified names. Each one was supposed to have a unique qualified name. While I was able to guarantee uniqueness within datasets and within taxon identifiers, I wasn't able to guarantee that qualified names would be unique between datasets and taxon identifiers.
So, here's the new version (click to magnify):
 Again, white arrows indicate "is-a" relationships ("inheritance")—so a PhyloDefinition is a type of Definition, a Dataset is a type of Qualified object, etc. And black diamonds indicate "has-a" relationships ("composition")—so a TaxonIdentifier has one (and only one) Taxon, an Equation has at least two TaxonIdentifier objects, etc. (I've left out a few non-core classes, like BioFile and UserAccount.)
Again, white arrows indicate "is-a" relationships ("inheritance")—so a PhyloDefinition is a type of Definition, a Dataset is a type of Qualified object, etc. And black diamonds indicate "has-a" relationships ("composition")—so a TaxonIdentifier has one (and only one) Taxon, an Equation has at least two TaxonIdentifier objects, etc. (I've left out a few non-core classes, like BioFile and UserAccount.)
Brief discussions of each class:
Authority.—An authority can be a publication, a person, a bioinformatics file, a database, a specimen catalogue, etc. Each authority has a canonical name (e.g., "Yale Peabody Museum: Vertebrate Paleontology Collection") and an optional abbreviation (e.g., "YPM-VP").
AuthorityIdentifer.—One or more identifiers may be used to indicate an authority, each one associated with a unique URI. Examples:
TaxonIdentifier & Taxon.—Formerly called "signifiers", taxon identifiers are qualified objects that each refer to a taxon. Taxon identifiers may be scientific names, vernacular names, specimen identifiers, character state descriptions, etc. As with authorities, each taxon may have more than one identifier referring to it. For example, the following qualified names all refer to the same species: <urn:isbn:0853010064::Abeillia+abeillei>, <http://iucnredlist.org::species:142883>, and <http://iucnredlist.org::common_name:Eng:Emerald-chinned+Hummingbird>.
Label.—Authorities, datasets, and taxon identifiers are all labelled entities, possessing one label object. Each label has a name, an optional abbreviation, and a flag telling whether it should be italicized. Labels are merely cosmetic, and need not be unique. They are used as the targets of searches, using Hibernate Search.
Definition.—Each definition has one taxon identifier, and only one definition pertains to that taxon identifier. How do I accommodate differing definitions, then? I use a concept from the PhyloCode: conversion. Consider the name "Aves". Under the ICZN, it refers to a suprafamilial ranked taxon with no type. According to Sereno's TaxonSearch, it refers to a node-based clade including Archaeopteryx. According to Gauthier and de Queiroz (2001), it refers to a crown group. But instead of having multiples definitions for the same identifier, I consider each definition to define a different identifier, each indicating a (potentially) different taxon: <urn:isbn:0853010064::Aves>, <http://www.taxonsearch.org/Archive/stem-archosauria-1.0.php::Aves>, and <urn:bici:0912532572(200112)%3C7:FDFDCD%3E2.0.TX;2-H::Aves>, respectively. In cases of conversion, the definition also indicates the original identifier.
PhyloDefinition & RankDefinition.—These have not changed much, except that they now refer directly to their specifers and types, respectively. No more useless "Anchor" class.
Dataset.—Instead of storing a bunch of relations of unspecified type, each type of relation falls within its own set. I've also added optional ratios for converting weights in phylogenetic networks to generations and/or years.
Equation.—I almost called this "Synonymy". This is a new type of relation, which asserts that two or more identifiers refer to the same taxon.
Heredity & Inclusion.—Heredity was previously called "Parentage". The new nomenclature better reflects its real meaning, since it models ancestor-descendant relationships, not necessarily parent-child. These two classes are little changed, except that now they don't both descend from a useless Relation class, so their nomenclature can be clearer (predecessor and superset used to be "a"; successor and subset used to be "b").
This schema is much cleaner, and will make for a more efficient server-side. I've already implemented the entities, removed deprecated code, and updated the relevant code. After some hiccups with a Hibernate upgrade, unit tests are working again. The back-end should be complete fairly soon (pending some ideas about user accounts), and then it will be time to look at some massive refactorings for the front end!
Another big problem was the home-brewed keyword search system I had going. Synchronizing the keyword lists was becoming problematic, and I realized there are already perfectly good (better, even) tools out there such as Hibernate Search. That's a chief rule of programming: don't reinvent something that people smarter than you, with more time on their hands, have already invented.
After a clear, honest look at the contextual relations, I came to a realization: they should be in the client, not the back end. No need to bog down the server with computing definition applications when it can be done in the client. That simplified things a great deal.
Another thing I didn't really need was categories. They were basically an ad hoc form of class inheritance, e.g., a species name is a nomen, a nomenclatural code is a publication, etc. For a little while I considered implementing this as a class hierarchy, as I had in earlier versions. But, really, this is irrelevant data—Names on Nodes doesn't really need to know what category an identifier falls in.
Finally, I had another problem in the way datasets and taxon identifiers (=signifiers) used qualified names. Each one was supposed to have a unique qualified name. While I was able to guarantee uniqueness within datasets and within taxon identifiers, I wasn't able to guarantee that qualified names would be unique between datasets and taxon identifiers.
So, here's the new version (click to magnify):
 Again, white arrows indicate "is-a" relationships ("inheritance")—so a PhyloDefinition is a type of Definition, a Dataset is a type of Qualified object, etc. And black diamonds indicate "has-a" relationships ("composition")—so a TaxonIdentifier has one (and only one) Taxon, an Equation has at least two TaxonIdentifier objects, etc. (I've left out a few non-core classes, like BioFile and UserAccount.)
Again, white arrows indicate "is-a" relationships ("inheritance")—so a PhyloDefinition is a type of Definition, a Dataset is a type of Qualified object, etc. And black diamonds indicate "has-a" relationships ("composition")—so a TaxonIdentifier has one (and only one) Taxon, an Equation has at least two TaxonIdentifier objects, etc. (I've left out a few non-core classes, like BioFile and UserAccount.)Brief discussions of each class:
Authority.—An authority can be a publication, a person, a bioinformatics file, a database, a specimen catalogue, etc. Each authority has a canonical name (e.g., "Yale Peabody Museum: Vertebrate Paleontology Collection") and an optional abbreviation (e.g., "YPM-VP").
AuthorityIdentifer.—One or more identifiers may be used to indicate an authority, each one associated with a unique URI. Examples:
- <urn:isbn:0853010064> (The International Code of Zoological Nomenclature, 4th Edition)
- <http://iczn.org/iczn> (Another way of referring to the ICZN.)
- <mailto:keesey@gmail.com> (myself)
- <http://peabody.yale.edu/collections/vp> (Yale Peabody Museum: Vertebrate Paleontology Collection)
- <urn:sha1:bc0ccc8a379edc44cf91b013d2da6238d4258a56> (a bioinformatics file, indicated by its SHA-1 hash key)
TaxonIdentifier & Taxon.—Formerly called "signifiers", taxon identifiers are qualified objects that each refer to a taxon. Taxon identifiers may be scientific names, vernacular names, specimen identifiers, character state descriptions, etc. As with authorities, each taxon may have more than one identifier referring to it. For example, the following qualified names all refer to the same species: <urn:isbn:0853010064::Abeillia+abeillei>, <http://iucnredlist.org::species:142883>, and <http://iucnredlist.org::common_name:Eng:Emerald-chinned+Hummingbird>.
Label.—Authorities, datasets, and taxon identifiers are all labelled entities, possessing one label object. Each label has a name, an optional abbreviation, and a flag telling whether it should be italicized. Labels are merely cosmetic, and need not be unique. They are used as the targets of searches, using Hibernate Search.
Definition.—Each definition has one taxon identifier, and only one definition pertains to that taxon identifier. How do I accommodate differing definitions, then? I use a concept from the PhyloCode: conversion. Consider the name "Aves". Under the ICZN, it refers to a suprafamilial ranked taxon with no type. According to Sereno's TaxonSearch, it refers to a node-based clade including Archaeopteryx. According to Gauthier and de Queiroz (2001), it refers to a crown group. But instead of having multiples definitions for the same identifier, I consider each definition to define a different identifier, each indicating a (potentially) different taxon: <urn:isbn:0853010064::Aves>, <http://www.taxonsearch.org/Archive/stem-archosauria-1.0.php::Aves>, and <urn:bici:0912532572(200112)%3C7:FDFDCD%3E2.0.TX;2-H::Aves>, respectively. In cases of conversion, the definition also indicates the original identifier.
PhyloDefinition & RankDefinition.—These have not changed much, except that they now refer directly to their specifers and types, respectively. No more useless "Anchor" class.
Dataset.—Instead of storing a bunch of relations of unspecified type, each type of relation falls within its own set. I've also added optional ratios for converting weights in phylogenetic networks to generations and/or years.
Equation.—I almost called this "Synonymy". This is a new type of relation, which asserts that two or more identifiers refer to the same taxon.
Heredity & Inclusion.—Heredity was previously called "Parentage". The new nomenclature better reflects its real meaning, since it models ancestor-descendant relationships, not necessarily parent-child. These two classes are little changed, except that now they don't both descend from a useless Relation class, so their nomenclature can be clearer (predecessor and superset used to be "a"; successor and subset used to be "b").
This schema is much cleaner, and will make for a more efficient server-side. I've already implemented the entities, removed deprecated code, and updated the relevant code. After some hiccups with a Hibernate upgrade, unit tests are working again. The back-end should be complete fairly soon (pending some ideas about user accounts), and then it will be time to look at some massive refactorings for the front end!
Labels:
databases,
definition,
Java,
Names on Nodes,
nomenclature,
ontology,
refactoring,
systematics,
web applications
03 March 2009
Refactoring "Names on Nodes" Entities, Part I
(Warning: If you are not me, this post may not make much sense. Same could be said for many recent posts. Sorry for all the self-indulgence here, lately, but I'm trying to work through a lot of thorny issues.)
Last year I wrote a post about some revisions to the entity schema of Names on Nodes, my longstanding project to automate the application of phylogenetic nomenclature. The revisions were pretty hefty, and necessitated a rewrite of much of the project. I got pretty far without making any further major modifications to the schema. But, after a few months of work, some flaws are beginning to show.
Once again, here is the UML diagram:
 And, once again: The white arrows indicate inheritance, i.e., "is-a" relationships. For example, a PhyloDefinition is a type of Definition. The black diamonds indicate composition, i.e., "has" relationships. For example, a Definition has any number of Anchor entities, each of which has exactly one Signifier entity.
And, once again: The white arrows indicate inheritance, i.e., "is-a" relationships. For example, a PhyloDefinition is a type of Definition. The black diamonds indicate composition, i.e., "has" relationships. For example, a Definition has any number of Anchor entities, each of which has exactly one Signifier entity.
So, the problems...
The nomenclature is confusing.
Not all of it, but some. What I was calling a SignifierIdentity is, in fact, a taxon (in a somewhat loose sense, i.e., any set of organisms, or subset of life, or whatever—more here), and a Signifier is just a taxon identifier. What I was calling an Authority is actually a authority identifier, and what I was calling an AuthorityIdentity ... is really an authority!
Anchors are insufficient.
The idea of the Anchor class was to allow every definition, be it rank-based or phylogenetic, to be connected with any number of taxa, namely, those taxa required by the definition. Each Anchor object specifies a taxon (through an identifier/signifier) and tells whether it is internal or external. I had hoped that this would work equally well for both rank-based and phylogenetic definitions, modeling biological types for the former and the specifiers for the latter. But there are some crucial differences between types and specifiers:
Why do Parentage and Inclusion both extend Relation? Because they can. They both require two ordered operands (parent and child for the former, superset and subset for the latter). There really is no other reason; modeling them this way doesn't make calculations faster (in fact, it slows them down), and gives no benefit otherwise. Furthermore, the Relation class is incapable of modeling other types of relations, like equation (i.e., subjective and heterodefinitional synonymy), which has two or more unordered operands. (Note: objective/homodefinitional synonymy is already well-handled by the relation of identifiers/signifiers to taxa/identities.)
Relators are insufficient.
Why do Definition, DefinitionApplication, and Dataset all extend Relator? Good question. The idea was that all of them indicate relations of some kind. But this resemblance only goes so far.
For a while, I had been considering taxonomic names as defined by different authorities to share the same identity. This proved unworkable. Instead, whenever an authority defines a name, it is either coining that name anew, or converting it into a new name (that happens to have the same spelling, but a different authority). For example, Aves under Linnaeus 1758 and Aves under the ICZN are the same thing, but Aves sensu Gauthier & de Queiroz 2001 and Aves sensu Sereno 2005 are different entities.
For this reason, a definition can be considered to have the same authority as the name it defines. Keeping an extra reference to an authority is redundant. Under this system, every name gets only one definition (if that).
Looking up contextual relations is awkward.
Those other problems are pretty minor compared to this one. One of the core ideas of Names on Nodes is that you are free to create a phylogenetic context. A context is basically a way of saying which datasets you want to use (and which you want to ignore). Every definition is true for all contexts, but the application of each definition may differ.
Thus, when looking up things like whether taxon A is ancestral to taxon B (something you have to do a lot when applying phylogenetic definitions), the algorithm has to look at every single relation and decide whether it belongs or not. Does it belong to a definition? Then it belongs. Does it belong to a definition application? Then it belongs if tat application is under the specified context. Does it belong to a dataset? Then it belongs if that dataset is included in the context. I optimized this a lot, but, at the end of the day, I was making it do something it did not really need to do. Which brings me to my last point.
Looking up contextual relations is not easily optimized.
The Context class is pretty bare bones, and that's not a good thing. I've been looking into implementing some of the optimizations present in Bender & al. 2005, but it's not possible with the current schema.
So, some revisions are needed. Not nearly as major as last time, but fairly significant. More in Part II, coming some day....
Last year I wrote a post about some revisions to the entity schema of Names on Nodes, my longstanding project to automate the application of phylogenetic nomenclature. The revisions were pretty hefty, and necessitated a rewrite of much of the project. I got pretty far without making any further major modifications to the schema. But, after a few months of work, some flaws are beginning to show.
Once again, here is the UML diagram:
 And, once again: The white arrows indicate inheritance, i.e., "is-a" relationships. For example, a PhyloDefinition is a type of Definition. The black diamonds indicate composition, i.e., "has" relationships. For example, a Definition has any number of Anchor entities, each of which has exactly one Signifier entity.
And, once again: The white arrows indicate inheritance, i.e., "is-a" relationships. For example, a PhyloDefinition is a type of Definition. The black diamonds indicate composition, i.e., "has" relationships. For example, a Definition has any number of Anchor entities, each of which has exactly one Signifier entity.So, the problems...
The nomenclature is confusing.
Not all of it, but some. What I was calling a SignifierIdentity is, in fact, a taxon (in a somewhat loose sense, i.e., any set of organisms, or subset of life, or whatever—more here), and a Signifier is just a taxon identifier. What I was calling an Authority is actually a authority identifier, and what I was calling an AuthorityIdentity ... is really an authority!
Anchors are insufficient.
The idea of the Anchor class was to allow every definition, be it rank-based or phylogenetic, to be connected with any number of taxa, namely, those taxa required by the definition. Each Anchor object specifies a taxon (through an identifier/signifier) and tells whether it is internal or external. I had hoped that this would work equally well for both rank-based and phylogenetic definitions, modeling biological types for the former and the specifiers for the latter. But there are some crucial differences between types and specifiers:
- A rank-based definition may not have a type; but a phylogenetic definition must have at least one specifier (usually two or more, but in theory you could get by with one, e.g., "Homo erectus (Dubois 1892) and all of its descendants," not that I'd recommend it in most cases).
- A specifier can be a character state description, but a type cannot. (Both can be taxonomic names or specimen identifiers).
- Types are always internal, so it's pointless to have to mark them as such.
- A type is always included in the taxon. A specifier, even an internal one, may not be, since phylogenetically-defined taxa are potentially empty.
Why do Parentage and Inclusion both extend Relation? Because they can. They both require two ordered operands (parent and child for the former, superset and subset for the latter). There really is no other reason; modeling them this way doesn't make calculations faster (in fact, it slows them down), and gives no benefit otherwise. Furthermore, the Relation class is incapable of modeling other types of relations, like equation (i.e., subjective and heterodefinitional synonymy), which has two or more unordered operands. (Note: objective/homodefinitional synonymy is already well-handled by the relation of identifiers/signifiers to taxa/identities.)
Relators are insufficient.
Why do Definition, DefinitionApplication, and Dataset all extend Relator? Good question. The idea was that all of them indicate relations of some kind. But this resemblance only goes so far.
- Rank-based definitions do indicate that the types are included by the defined identifier/signifier, but phylogenetic definitions don't really indicate anything, since they potentially yield empty results.
- The inclusions indicated by rank-based definitions are redundant with the information about their types. I had to implement an awkward system to synchronize this.
- Datasets are the only relators that can indicate parentage; the other two can only indicate inclusion.
- Only datasets can indicate subjective synonymy, and only definition applications can indicate heterodefinitional synonymy. Those relations aren't currently modeled at all, but should be.
For a while, I had been considering taxonomic names as defined by different authorities to share the same identity. This proved unworkable. Instead, whenever an authority defines a name, it is either coining that name anew, or converting it into a new name (that happens to have the same spelling, but a different authority). For example, Aves under Linnaeus 1758 and Aves under the ICZN are the same thing, but Aves sensu Gauthier & de Queiroz 2001 and Aves sensu Sereno 2005 are different entities.
For this reason, a definition can be considered to have the same authority as the name it defines. Keeping an extra reference to an authority is redundant. Under this system, every name gets only one definition (if that).
Looking up contextual relations is awkward.
Those other problems are pretty minor compared to this one. One of the core ideas of Names on Nodes is that you are free to create a phylogenetic context. A context is basically a way of saying which datasets you want to use (and which you want to ignore). Every definition is true for all contexts, but the application of each definition may differ.
Thus, when looking up things like whether taxon A is ancestral to taxon B (something you have to do a lot when applying phylogenetic definitions), the algorithm has to look at every single relation and decide whether it belongs or not. Does it belong to a definition? Then it belongs. Does it belong to a definition application? Then it belongs if tat application is under the specified context. Does it belong to a dataset? Then it belongs if that dataset is included in the context. I optimized this a lot, but, at the end of the day, I was making it do something it did not really need to do. Which brings me to my last point.
Looking up contextual relations is not easily optimized.
The Context class is pretty bare bones, and that's not a good thing. I've been looking into implementing some of the optimizations present in Bender & al. 2005, but it's not possible with the current schema.
So, some revisions are needed. Not nearly as major as last time, but fairly significant. More in Part II, coming some day....
Labels:
definition,
Names on Nodes,
nomenclature,
ontology,
OOP,
phylogeny
23 February 2009
Brand New Forum for Discussion of Phylogenetic Nomenclature
The title says it all. As some of you may know, the ISPN's online forum has been down for a while. Daniel Madzia, of Wild Prehistory, has taken it upon himself to create a new forum: PhyloNom.

We've started a few threads. If you are interested in biological nomenclature, come on over and check it out!

We've started a few threads. If you are interested in biological nomenclature, come on over and check it out!
Labels:
definition,
ispn,
nomenclature,
PhyloCode,
phylogeny,
taxonomy
13 February 2009
Using Conservation Status to Automatically Apply Phylogenetic Definitions
To briefly summarize some relevant points in the last post (Extinct or Extant?):
- Some phylogenetic definitions require a definition of the term "extant".
- The International Union for the Conservation of Nature maintains a database of species and their conservation status, as assessed for a particular year.
EX: ExtinctEW: Extinct in the WildCR: Critically EndangeredEN: EndangeredVU: VulnerableNT: Near ThreatenedLC: Least ConcernDD: Data DeficientNE: Not Evaluated
urn:isbn:2831706335::categories:EX:2008urn:isbn:2831706335::categories:CR:2008urn:isbn:2831706335::categories:EN:2008urn:isbn:2831706335::categories:VU:2008urn:isbn:2831706335::categories:NT:2008urn:isbn:2831706335::categories:LC:2008urn:isbn:2831706335::categories:DD:2008urn:isbn:2831706335::categories:NE:2008
<species id="148296">This provides a source not only for the conservation status of species, but also for the species themselves and some of their higher taxa as well. This one XML snippet can provide all of the following signifiers:
<scientific_name>
Zosterops xanthochroa
</scientific_name>
<kingdom_name>
ANIMALIA
</kingdom_name>
<phylum_name>
CHORDATA
</phylum_name>
<class_name>
AVES
</class_name>
<order_name>
Passeriformes
</order_name>
<family_name>
Zosteropidae
</family_name>
<genus_name>
Zosterops
</genus_name>
<species_name>
xanthochroa
</species_name>
<authority>
Gray, 1859
</authority>
<synonyms>
<synonym>
<scientific_name>
Zosterops xanthochrous
</scientific_name>
<genus_name>
Zosterops
</genus_name>
<species_name>
xanthochrous
</species_name>
</synonym>
</synonyms>
<common_names>
<name lang="Eng">
Green-backed White-eye
</name>
</common_names>
<assessment
version="3.1"
year="2008">
<category>
LC
</category>
</assessment>
</species>
- Animalia
urn:isbn:0853010064::Animalia
- Chordata
urn:isbn:0853010064::Chordata
- Aves
urn:isbn:0853010064::Aves
- Passeriformes
urn:isbn:0853010064::Passeriformes
- Zosteropoidea
urn:isbn:0853010064::Zosteropoidea
- Zosteropidae
urn:isbn:0853010064::Zosteropidae
- Zosteropinae
urn:isbn:0853010064::Zosteropinae
- Zosteropini
urn:isbn:0853010064::Zosteropini
- Zosteropina
urn:isbn:0853010064::Zosteropina
- Zosterops
urn:isbn:0853010064::Zosterops
- Zosterops (Zosterops)
urn:isbn:0853010064::Zosterops+%28Zosterops%29
- Zosterops xanthochroa/Zosterops xanthochrous/Green-backed White-eye
urn:isbn:0853010064::Zosterops+xanthochroaurn:isbn:0853010064::Zosterops+xanthochroushttp://iucnredlist.org::species:148296http://iucnredlist.org::common_name:Eng:Green-backed+White-eye
<apply xmlns="http://www.w3.org/1998/Math/MathML">Presto, now I can apply modified node-based definitions and total group definitions! Thanks, IUCN, for helping to enable the automated application of phylogenetic definitions! (And, you know, also for all the "saving threatened species from extinction" stuff.)
<union/>
<csymbol
definitionURL="urn:isbn:2831706335::categories:EW:2008"/>
<csymbol
definitionURL="urn:isbn:2831706335::categories:CR:2008"/>
<csymbol
definitionURL="urn:isbn:2831706335::categories:EN:2008"/>
<csymbol
definitionURL="urn:isbn:2831706335::categories:VU:2008"/>
<csymbol
definitionURL="urn:isbn:2831706335::categories:NT:2008"/>
<csymbol
definitionURL="urn:isbn:2831706335::categories:LC:2008"/>
</apply>
Labels:
conservation,
databases,
definition,
MathML,
Names on Nodes,
nomenclature,
phylogeny
12 February 2009
Extinct or Extant?
It's pretty easy to tell whether something's alive, right? You might have to jab it with a stick a couple of times to make sure (assuming it's an animal), but generally it's not too hard. So you'd think.
The International Union for Conservation of Nature is devoted to the preservation of life's diversity, so, naturally, it has a big stake in this question. When should we expend energy to try to save a critically endangered species, and when should we throw in the towel? Their Red List Guidelines say this about extinction:
It would be nice if we could simply categorize species as extinct or extant, but it's not always easy. A species may be extant one day and extinct the next. And we may not realize this until years or even decades later. Or, we may think a species extinct only to have individuals turn up again, as may have happened with Campephilus principalis, the ivory-billed woodpecker, a few years ago (Hill et al. 2006).
This question is important not only for conservation efforts, but also for nomenclature. In fact, in some ways, the issues become even thornier for nomenclature. To see why this is so, let's look at the phylogenies of two mammalian taxa.
Cetacea is what we call a "crown group". A crown group is a special type of clade, a clade being an ancestor and all of its descendants. A crown group is the final common ancestor of certain extant organisms, and all descendants of that ancestor. Note that this doesn't mean that all members of a crown group are extant; for example, Aetiocetus, a proto-baleen whale known from fossils, is long extinct. But it is descended from the final common ancestor of living baleen whales (Mysticeti) and living toothed whales (Odontoceti), so it is still a member of the crown group Cetacea.
The cetacean "total group", informally termed "pan-Cetacea", includes everything sharing closer ancestry with cetaceans than with any other extant organisms. A number of extinct taxa, from Pakicetus to Dorudon, are members of the total group, but not of the crown group. Therefore, they are part of the cetacean "stem group", or, more succinctly, "stem-cetaceans". (Indohyus may also be a stem-cetacean, but there are differing hypotheses.) Note that the stem group includes the ancestors of the crown group, but not all members of the stem group are ancestors of the crown group. For example, Basilosaurus cetoides is a stem-cetacean, but it is a somewhat derived offshoot of the cetacean lineage, with a long, snake-like body different from that of modern cetaceans or their ancestors.
Somewhere around the time of the Cretaceous-Paleogene extinction (when non-avian dinosaurs, among many other taxa, became extinct reached a population size of zero), the cetacean line split off from other extant lineages (either from the hippopotamid lineage, the ruminant lineage, or both at once—the artiodactyl lineage). The earliest stem-cetaceans were hoofed, but they soon gave way to amphibious varieties, which looked vaguely like mammalian crocodiles with flippers. Over time, adaptations toward an aquatic lifestyle were accumulated in stem-cetacean populations: tail flukes, dorsal fins, birth in the water. Stem-cetaceans were replaced by cetaceans, which possessed all of these adaptations. Early cetaceans split into two major lineages: one leading to the filter-feeding mysticetes and the other to the echolocating, predatory odontocetes.
Many living species of cetacean are threatened. Perhaps the worst case is that of the Yangtze River dolphin or baiji, Lipotes vexillifer. This human-sized freshwater cetacean was once one of the few animals to be actually protected by superstition (many others, instead, are endangered by it—think of rhinoceros horns as an ingredient in impotence remedies). But, in modern times, this protection has come to mean less. The last uncontested sighting of a baiji was in 2004. The IUCN currently classifies the species as critically endangered, but it may be extinct already. If so, it would be the first aquatic mammal species to go extinct in the 3rd millennium—less than a decade in and we're already off to a bad start.
Not all zoologists use Cetacea in the crown group sense; some paleontologists expand it to include some or all of the stem group. But there is a danger in doing this. Cetacea is primarily a term from the neontological (as opposed to paleontological) literature, so it is most often associated with the suite of characters that the living organisms possess. But members of the stem group may or may not possess these. A recent, spectacular discovery of a fossilized, pregnant Maiacetus (which would go in the above chart somewhere around Rodhocetus) shows that Maiacetus probably gave birth on land. It is not known (to me, anyway) whether they had dorsal fins or tail flukes.
Since extending neontological terms beyond the crown group can result in unwarranted character inferences, some systematists prefer to limit such terms to crown groups when possible. The PhyloCode, a nomenclatural code currently in draft form, advocates this approach (see, for example Recommendation 10.1B). (The PhyloCode is also the source of the "pan-" convention for the names of total groups; see Art. 10.3.)
Moving too fast? Let's slow down....
Here is a phylogeny with a sampling of species to give an overview of sloth evolution (again, highly abridged, to say the least):
If you look at the above diagram, you might think, "But, look, there are more than just two extant groups." This is because the diagram is on such a vast scale that it's impossible to distinguish the extant from the recently extinct. Here's the same phylogeny to a logarithmic scale, which expands recent time:

Now we can actually see the Holocene, or "Recent", our current geological epoch (unless you accept the Anthropocene—more on that later). And you can see that some taxa, such as Mylodon and Megatherium, died out around the Pleistocene-Holocene transition. This transition was only 11 to 12 thousand years ago (an eyeblink in geological time, as can be seen by the fact that it's not even visible in the first chart).
Some Haitian sloth species persisted until much more recent times. Parocnus serus and Synocnus comes were still hanging around (ha ha—just kidding, they were more or less terrestrial) when European explorers first came to the Caribbean. They may have died out in the 16th century C.E.
Sloths present an interesting case because the clades that can be considered crown groups have changed over the course of human existence. Twelve thousand years ago, when humans were still settling the New World, a sloth crown group would have included Mylodon, and within that group a smaller crown group would have included Choloepus, Hapalops, Thalassocnus, Megatherium, Synocnus, and Parocnus. (Thalassocnus and Hapalops were extinct, but would still be part of that crown group.) After the Holocene-Pleistocene extinctions, Mylodon would no longer be part of the sloth crown group, and the Choloepus-but-not-Bradypus crown group would no longer contain Thalassocnus, Megatherium, or Hapalops. This continued, more or less, until the European/African settling of the Caribbean, at which time Synocnus and Parocnus died out.
Today, some species of Bradypus (B. pygmaeus and B. torquatus) are endangered. Time will tell if conservation efforts win out, or if the Bradypus crown group shrinks further.
There are three major ways to define a crown group:
There is a peril with this approach, though: what if a new phylogenetic hypothesis shows some member to be outside the delimited clade? Fortunately the PhyloCode allows for expedient "unrestricted" emendations in such cases (i.e., minor, commonsense emendations that don't require committe approval; see Art. 15). But ideally the need for such emendations should be avoided. One way to avoid this need is with modified node-based definitions, which come in two major flavors.
There are two pitfalls to this approach. One is that you might fail to specify the closest extant outgroup. For example, if pigs (suids) turned out to be closer to whales than cattle or hippos are, then, under that definition, pigs would be cetaceans! Again, this can be fixed with an unrestricted emendation, but it would be nice not to have to do that.
The other pitfall is that the author(s) must define "extant", but more on that later.
There are two pitfalls with this approach. One is that the apomorphy may turn out not to have evolved within the stem group. It may have evolved earlier, thus expanding the content of the clade, or it may have evolved independently multiple times within the crown group, thus contracting the content of the clade. (It must be said, though, that in the case of cetacean tail flukes, both possibilities are extremely unlikely.)
The other pitfall is the same as that of branch-modified node-based definitions: what does "extant" mean? Extant when? And by what criteria? Let's look at this in more depth.
I find this problematic for a couple of reasons. One is that the Holocene covers all of human history and more, so just being Holocene is no guarantee that we'll have good specimens. Some Holocene species went extinct thousands of years before Sumerians ever put wedge to clay tablet. Look at the sloth phylogeny—some of the species, such as Mylodon sp. and M. americanum, seem to have gone extinct right before the Holocene. But what if some small populations endured for a short while in refugia? That could drastically change the content of, e.g., a branch-modified node-based clade including Choloepus but not Bradypus.
The other problem is that "Recent" doesn't really get at the reason why crown groups are interesting. They're interesting because we have a wealth of available data about some of their members, data which can be used to extrapolate ancestral states. The same amount of data is not present for stem groups, which are generally known from fossils, if they are known at all.
This approach also gets closer to the basic intent of crown groups. Extra data are available in non-fossil specimens. But it's still short of the data present in living forms; for example, behavior is not observable. Is it enough extra data to warrant recognizing the species as extant for nomenclatural purposes? It boils down to opinion. (And I note that behavior might not be a very important consideration for Phil's purposes, since he works on plants.)
This idea has direct relevance for sloths, because one extinct form is actually represented by non-fossil specimens! Mylodon skins, complete with armor nodules and fur, still exist, having been preserved in caves. Supposed that Folivora were defined as the clade originating with the final common ancestor of all extant organisms sharing closer ancestry with Bradypus tridactylus Linnaeus 1758 than with Myrmecophaga tridactyla Linnaeus 1758 (the giant anteater). The question of whether Mylodon is extant would determine whether an entire clade (Mylodontidae) belongs to Folivora. (Of course, nobody says that has to be the definition of Folivora, or even that Folivora has to be a crown group, but this is just an example.)
This designation hasn't met widespread adoption, to my knowledge, nor has it been proposed as a criterion for determining whether a species is "extant" for the purposes of nomenclature. But it seems to me like a better candidate than the Holocene. At least Anthropocene species have all coexisted with scientists.
Both of these approaches (Anthropocene and Systema Naturae) have similar problems to the use of "Recent", although to a lesser extent. It's difficult to establish whether some species went extinct before or after the selected boundary. For example, the sloths Synocnus and Parocnus probably went extinct a couple of centuries earlier than these dates, but it's possible that they persisted in remote areas. An even closer example is Hydrodamalis gigas, Steller's sea cow, which seems to have gone extinct by 1768 (post-Systema Naturae, pre-Anthropocene!).
Well, there is no one "now". Every instant is its own "now". Obviously, I mean something closer to the PhyloCode's default definition: extant as of the publication date of the definition.
This is less problematic than using earlier dates in some ways. We have much better ways of tracking populations today than we did in the 1700s. But pushing the date closer to the present also presents problems. Consider Steller's sea cow and the Yangtze River dolphin. It's easy to say that the sea cow is extinct, but the fate of the dolphin is still as unclear as the muddy waters it swims (or swam?) in. Consider: what if, despite the phylogeny presented above, Lipotes was found to be an outgroup to [other] extant cetaceans? Would the cetacean crown group include it or not? (Thanks to Matt Martyniuk for thinking of that example.)
And all of the meanings mentioned so far share another problem: the discovery of a previously unknown species could change everything. There are many example of "Lazarus taxa" (so-called because, like the character of Lazarus in the Christian gospels, they appear to rise from the grave), living organisms that represent clades previously known only from fossils: the Laotian rock rat, Laonastes aenigmamus (Diatomyidae); the Indian Ocean coelacanth, Latimeria; the gladiators, Mantophasmatinae (Insecta: Mantophasmatodea); the monito del monte, Dromiciops gliroides (Marsupialia: Microbiotheria); the Wollemi pine, Wollemia nobilis (Araucariaceae: Wollemia); etc. Although the discovery of such a species is always a wonderful event, it's potentially disruptive to modified node-based definitions.
But this still doesn't solve the problem of whether Lipotes is extinct or extant.
Of course, the IUCN doesn't cover all species, leaving out 1) species that have been extinct for a long time (e.g., Tyrannosaurus rex), and 2) species that haven't been published by scientists yet (e.g., Laonastes aenigmamus in lists prior to 2005). But I think in both of these cases we can consider such species to be "non-extant for the purposes of nomenclature". Long-extinct species are clearly not extant. Treating undiscovered species as non-extant has the same stabilizing benefit as requiring an extant species to be published upon. The only problem spot is the taxa that the IUCN doesn't focus on, e.g., bacteria and archaeans. But this still leaves plenty of taxa that it works just fine for.
I think I like this approach best, at least for the taxa I study (amniotes). Delegate the issue to the experts. Mylodon and Synocnus are extinct. Lipotes is critically endangered (at least as of last year). The nomenclatural problem is taken care of, and we can move on to more crucial problems, like preserving the crown groups that we have.
The International Union for Conservation of Nature is devoted to the preservation of life's diversity, so, naturally, it has a big stake in this question. When should we expend energy to try to save a critically endangered species, and when should we throw in the towel? Their Red List Guidelines say this about extinction:
Extinction is defined as population size reaching zero.That made me laugh when I first read it. Really? You don't say! But I read on, and it became clear that there was much more to this seemingly simple definition:
Population size is the number of all individuals of the taxon (not only mature individuals). In some cases, extinction can be defined as population size reaching a number larger than zero. For example, if only females are modelled, it is prudent to define extinction as one female (instead of zero) remaining in the population. More generally, an extinction threshold greater than zero is justified if factors that were not incorporated into the analysis due to a lack of information (for example, Allee effects, sex structure, genetics, or social interactions) make the predictions of the analysis at low population sizes unreliable.This is just a small sample of what the IUCN has to say on the subject. So much for just poking things with sticks.
For Criterion E, extinction risk must be calculated for up to 3 different time periods:
For a taxon with a generation length of 34 years or longer, only one assessment for 100 years) is needed. For a taxon with a generation length of 20 to 33 years, two assessments (for 3 generations and 100 years) are needed. For a taxon with a generation length less than 20 years, all three assessments are needed.
- 10 years or 3 generations, whichever is longer (up to a maximum of 100 years)
- 20 years or 5 generations, whichever is longer (up to a maximum of 100 years)
- 100 years
It would be nice if we could simply categorize species as extinct or extant, but it's not always easy. A species may be extant one day and extinct the next. And we may not realize this until years or even decades later. Or, we may think a species extinct only to have individuals turn up again, as may have happened with Campephilus principalis, the ivory-billed woodpecker, a few years ago (Hill et al. 2006).
This question is important not only for conservation efforts, but also for nomenclature. In fact, in some ways, the issues become even thornier for nomenclature. To see why this is so, let's look at the phylogenies of two mammalian taxa.
Whales
Whales, or cetaceans, are related to even-toed ungulates, or artiodactyls. (In fact, they may even be artiodactyls, but that's a discussion that I'm going to try to avoid as much as possible right now.) The chart below shows a sampling of fossil and living species, giving a very rough and highly abridged picture of cetacean evolution:Time goes from left to right. Arrows point from ancestor species to descendant species. Silhouettes are not to scale.
Cetacea is what we call a "crown group". A crown group is a special type of clade, a clade being an ancestor and all of its descendants. A crown group is the final common ancestor of certain extant organisms, and all descendants of that ancestor. Note that this doesn't mean that all members of a crown group are extant; for example, Aetiocetus, a proto-baleen whale known from fossils, is long extinct. But it is descended from the final common ancestor of living baleen whales (Mysticeti) and living toothed whales (Odontoceti), so it is still a member of the crown group Cetacea.
The cetacean "total group", informally termed "pan-Cetacea", includes everything sharing closer ancestry with cetaceans than with any other extant organisms. A number of extinct taxa, from Pakicetus to Dorudon, are members of the total group, but not of the crown group. Therefore, they are part of the cetacean "stem group", or, more succinctly, "stem-cetaceans". (Indohyus may also be a stem-cetacean, but there are differing hypotheses.) Note that the stem group includes the ancestors of the crown group, but not all members of the stem group are ancestors of the crown group. For example, Basilosaurus cetoides is a stem-cetacean, but it is a somewhat derived offshoot of the cetacean lineage, with a long, snake-like body different from that of modern cetaceans or their ancestors.
Somewhere around the time of the Cretaceous-Paleogene extinction (when non-avian dinosaurs, among many other taxa, became extinct reached a population size of zero), the cetacean line split off from other extant lineages (either from the hippopotamid lineage, the ruminant lineage, or both at once—the artiodactyl lineage). The earliest stem-cetaceans were hoofed, but they soon gave way to amphibious varieties, which looked vaguely like mammalian crocodiles with flippers. Over time, adaptations toward an aquatic lifestyle were accumulated in stem-cetacean populations: tail flukes, dorsal fins, birth in the water. Stem-cetaceans were replaced by cetaceans, which possessed all of these adaptations. Early cetaceans split into two major lineages: one leading to the filter-feeding mysticetes and the other to the echolocating, predatory odontocetes.
Many living species of cetacean are threatened. Perhaps the worst case is that of the Yangtze River dolphin or baiji, Lipotes vexillifer. This human-sized freshwater cetacean was once one of the few animals to be actually protected by superstition (many others, instead, are endangered by it—think of rhinoceros horns as an ingredient in impotence remedies). But, in modern times, this protection has come to mean less. The last uncontested sighting of a baiji was in 2004. The IUCN currently classifies the species as critically endangered, but it may be extinct already. If so, it would be the first aquatic mammal species to go extinct in the 3rd millennium—less than a decade in and we're already off to a bad start.
Not all zoologists use Cetacea in the crown group sense; some paleontologists expand it to include some or all of the stem group. But there is a danger in doing this. Cetacea is primarily a term from the neontological (as opposed to paleontological) literature, so it is most often associated with the suite of characters that the living organisms possess. But members of the stem group may or may not possess these. A recent, spectacular discovery of a fossilized, pregnant Maiacetus (which would go in the above chart somewhere around Rodhocetus) shows that Maiacetus probably gave birth on land. It is not known (to me, anyway) whether they had dorsal fins or tail flukes.
Since extending neontological terms beyond the crown group can result in unwarranted character inferences, some systematists prefer to limit such terms to crown groups when possible. The PhyloCode, a nomenclatural code currently in draft form, advocates this approach (see, for example Recommendation 10.1B). (The PhyloCode is also the source of the "pan-" convention for the names of total groups; see Art. 10.3.)
Moving too fast? Let's slow down....
Sloths
Although today's sloths, or Folivora ("leaf-eaters"), are tree-dwellers, many in the past were terrestrial; some were even amphibious (living sloths are good swimmers when they need to be). Modern sloths exist in two clades: Bradypus, the three-toed sloths, and Choloepus, the two-toed sloths. The closest living relatives to sloths are Vermilingua ("worm-tongues"), or "true" anteaters (not to be confused with other long-tongued mammals that feed on eusocial insects, such as aardvarks, numbats, and echidnas). Together, sloths and anteaters comprise a clade called Pilosa ("hairy ones"). All living pilosans are Neotropical, although some fossil taxa were Nearctic (as are some of their cousins, the armadillos, or Loricata).Here is a phylogeny with a sampling of species to give an overview of sloth evolution (again, highly abridged, to say the least):
The sloth lineage split from its stem-anteater kin during the Paleocene. The original sloths were terrestrial, but at least two clades became highly arboreal (Bradypus and Choloepus, mentioned before). One clade, including Thalassocnus, went in a different direction and became amphibious. Most lineages, however, remained terrestrial, one of them culminating in the enormous Megatherium americanum, a sloth the size of an elephant.Time goes from left to right. Left-right lines connect ancestor species to descendant species. Silhouettes are not to scale.
Note that I've flipped the living sloths upside-down ... err, right-side-up ... err ... never mind.
If you look at the above diagram, you might think, "But, look, there are more than just two extant groups." This is because the diagram is on such a vast scale that it's impossible to distinguish the extant from the recently extinct. Here's the same phylogeny to a logarithmic scale, which expands recent time:

Now we can actually see the Holocene, or "Recent", our current geological epoch (unless you accept the Anthropocene—more on that later). And you can see that some taxa, such as Mylodon and Megatherium, died out around the Pleistocene-Holocene transition. This transition was only 11 to 12 thousand years ago (an eyeblink in geological time, as can be seen by the fact that it's not even visible in the first chart).
Some Haitian sloth species persisted until much more recent times. Parocnus serus and Synocnus comes were still hanging around (ha ha—just kidding, they were more or less terrestrial) when European explorers first came to the Caribbean. They may have died out in the 16th century C.E.
Sloths present an interesting case because the clades that can be considered crown groups have changed over the course of human existence. Twelve thousand years ago, when humans were still settling the New World, a sloth crown group would have included Mylodon, and within that group a smaller crown group would have included Choloepus, Hapalops, Thalassocnus, Megatherium, Synocnus, and Parocnus. (Thalassocnus and Hapalops were extinct, but would still be part of that crown group.) After the Holocene-Pleistocene extinctions, Mylodon would no longer be part of the sloth crown group, and the Choloepus-but-not-Bradypus crown group would no longer contain Thalassocnus, Megatherium, or Hapalops. This continued, more or less, until the European/African settling of the Caribbean, at which time Synocnus and Parocnus died out.
Today, some species of Bradypus (B. pygmaeus and B. torquatus) are endangered. Time will tell if conservation efforts win out, or if the Bradypus crown group shrinks further.
Defining Crown Groups
I've been talking about crown groups changing over time, but we need nomenclature to be stable. (Why? Well, for one thing, so we can communicate effectively about conservation efforts.) One way to do this is to tie names to phylogeny-based definitions. This is how the PhyloCode works.There are three major ways to define a crown group:
1. Node-Based Definition
This is the simplest way: just build up a list of extant specifiers, take their final common ancestor, and add all descendants. As an example, we could define Cetacea as the clade originating with the final common ancestor of Balaena mysticetus Linnaeus 1758 and Delphinus phocaena Linnaeus 1758 (=Phocoena phocaena Gray 1825). One advantage of this type of definition is that we don't need to worry about the meaning of "extant".There is a peril with this approach, though: what if a new phylogenetic hypothesis shows some member to be outside the delimited clade? Fortunately the PhyloCode allows for expedient "unrestricted" emendations in such cases (i.e., minor, commonsense emendations that don't require committe approval; see Art. 15). But ideally the need for such emendations should be avoided. One way to avoid this need is with modified node-based definitions, which come in two major flavors.
2. Branch-Modified Node-Based Definition
In this approach, we create a node-based definition using all extant members of a given total group. For example, the cetacean total group could be defined as everything sharing closer ancestry with B. mysticetus than with Hippopotamus amphibius Linnaeus 1758 or Bos taurus Linnaeus 1758. Thus, Cetacea could be defined as the clade originating with the final common ancestor of all extant organisms that share a closer common ancestor with B. mysticetus than with H. amphibius or B. taurus.There are two pitfalls to this approach. One is that you might fail to specify the closest extant outgroup. For example, if pigs (suids) turned out to be closer to whales than cattle or hippos are, then, under that definition, pigs would be cetaceans! Again, this can be fixed with an unrestricted emendation, but it would be nice not to have to do that.
The other pitfall is that the author(s) must define "extant", but more on that later.
3. Apomorphy-Modified Node-Based Definition
This style of definition uses a derived character, or "apomorphy", to delimit a clade, and then creates a node-based clade using the members of that apomorphy-based clade. This requires some apomorphy that evolved within the stem group. Cetacea, for example, could be defined as the clade originating with the final common ancestor of all extant organisms that possess tail flukes homologous (synapomorphic) with those of B. mysticetus.There are two pitfalls with this approach. One is that the apomorphy may turn out not to have evolved within the stem group. It may have evolved earlier, thus expanding the content of the clade, or it may have evolved independently multiple times within the crown group, thus contracting the content of the clade. (It must be said, though, that in the case of cetacean tail flukes, both possibilities are extremely unlikely.)
The other pitfall is the same as that of branch-modified node-based definitions: what does "extant" mean? Extant when? And by what criteria? Let's look at this in more depth.
The Many Flavors of "Extant"
Although many of the PhyloCode's articles deal with crown groups and total groups, the code doesn't provide a single definition of "extant". Instead, the author of the definition must select a meaning. The author has considerable latitude here. If nothing is specified, there is a default fallback: extant at time of publication (Art. 9.5).Recent (Holocene)
In just about every place that the PhyloCode uses the word "extant", it is followed with a parenthesis: "(or Recent)". In other words, a crown group may be considered as a clade originating with the final common ancestor of Holocene organisms.I find this problematic for a couple of reasons. One is that the Holocene covers all of human history and more, so just being Holocene is no guarantee that we'll have good specimens. Some Holocene species went extinct thousands of years before Sumerians ever put wedge to clay tablet. Look at the sloth phylogeny—some of the species, such as Mylodon sp. and M. americanum, seem to have gone extinct right before the Holocene. But what if some small populations endured for a short while in refugia? That could drastically change the content of, e.g., a branch-modified node-based clade including Choloepus but not Bradypus.
The other problem is that "Recent" doesn't really get at the reason why crown groups are interesting. They're interesting because we have a wealth of available data about some of their members, data which can be used to extrapolate ancestral states. The same amount of data is not present for stem groups, which are generally known from fossils, if they are known at all.
Non-Fossil Specimens
Philip Cantino, one of the authors of the PhyloCode, once told me (pers. comm.) his opinion on what "extant" should mean: "I think that any species that was extant recently enough to be represented in museums in a non-fossilized form (e.g., study skins, herbarium specimens) should be treated as extant." Note one big advantage of this approach: it's much simpler to verify whether something is extant.This approach also gets closer to the basic intent of crown groups. Extra data are available in non-fossil specimens. But it's still short of the data present in living forms; for example, behavior is not observable. Is it enough extra data to warrant recognizing the species as extant for nomenclatural purposes? It boils down to opinion. (And I note that behavior might not be a very important consideration for Phil's purposes, since he works on plants.)
This idea has direct relevance for sloths, because one extinct form is actually represented by non-fossil specimens! Mylodon skins, complete with armor nodules and fur, still exist, having been preserved in caves. Supposed that Folivora were defined as the clade originating with the final common ancestor of all extant organisms sharing closer ancestry with Bradypus tridactylus Linnaeus 1758 than with Myrmecophaga tridactyla Linnaeus 1758 (the giant anteater). The question of whether Mylodon is extant would determine whether an entire clade (Mylodontidae) belongs to Folivora. (Of course, nobody says that has to be the definition of Folivora, or even that Folivora has to be a crown group, but this is just an example.)
Anthropocene
Although the Holocene is already a ridiculously short geological epoch, Cruzen and Stoermer (2000) proposed naming a new, much shorter geological epoch for the Industrial Age. They named the "Anthropocene" in recognition of the global effects that Industrial-Age humans have had upon the environment, and set its starting date as 1784 C.E., with James Watts' invention of the steam engine. (This is also, not coincidentally, around the time that certain effects of pollution start to appear in ice core samples.)This designation hasn't met widespread adoption, to my knowledge, nor has it been proposed as a criterion for determining whether a species is "extant" for the purposes of nomenclature. But it seems to me like a better candidate than the Holocene. At least Anthropocene species have all coexisted with scientists.
Living at a Given Time in History
A similar candidate to using the Anthropocene, was proposed in a bulletin board discussion by Mike Taylor. Under this proposal, anything living during or after 1758 C.E. would be considered extant, 1758 being the year that the 10th edition of Linnaeus' Systema Naturae was published. That publication is regarded as the beginning of biological nomenclature by the botanical and zoological codes.Both of these approaches (Anthropocene and Systema Naturae) have similar problems to the use of "Recent", although to a lesser extent. It's difficult to establish whether some species went extinct before or after the selected boundary. For example, the sloths Synocnus and Parocnus probably went extinct a couple of centuries earlier than these dates, but it's possible that they persisted in remote areas. An even closer example is Hydrodamalis gigas, Steller's sea cow, which seems to have gone extinct by 1768 (post-Systema Naturae, pre-Anthropocene!).
Living Now
Right now. Wait, I mean NOW. Wait ... no ... okay ... NOW.Well, there is no one "now". Every instant is its own "now". Obviously, I mean something closer to the PhyloCode's default definition: extant as of the publication date of the definition.
This is less problematic than using earlier dates in some ways. We have much better ways of tracking populations today than we did in the 1700s. But pushing the date closer to the present also presents problems. Consider Steller's sea cow and the Yangtze River dolphin. It's easy to say that the sea cow is extinct, but the fate of the dolphin is still as unclear as the muddy waters it swims (or swam?) in. Consider: what if, despite the phylogeny presented above, Lipotes was found to be an outgroup to [other] extant cetaceans? Would the cetacean crown group include it or not? (Thanks to Matt Martyniuk for thinking of that example.)
And all of the meanings mentioned so far share another problem: the discovery of a previously unknown species could change everything. There are many example of "Lazarus taxa" (so-called because, like the character of Lazarus in the Christian gospels, they appear to rise from the grave), living organisms that represent clades previously known only from fossils: the Laotian rock rat, Laonastes aenigmamus (Diatomyidae); the Indian Ocean coelacanth, Latimeria; the gladiators, Mantophasmatinae (Insecta: Mantophasmatodea); the monito del monte, Dromiciops gliroides (Marsupialia: Microbiotheria); the Wollemi pine, Wollemia nobilis (Araucariaceae: Wollemia); etc. Although the discovery of such a species is always a wonderful event, it's potentially disruptive to modified node-based definitions.
Living And Published Upon
This last problem can be easily remedied, though: just require that something must be extant and published upon at the time of the definition. This could go a long way toward stabilizing definitions. The only drawback is that it could be seen as a bit arrogant: "If science hasn't heard of it, then it doesn't exist!" But this is only for nomenclatural purposes (of course Wollemi pines make a sound when they fall, whether scientists hear it or not).But this still doesn't solve the problem of whether Lipotes is extinct or extant.
Let Someone Else Worry About It
The IUCN has put tons of thought and effort into these sorts of questions. One possibility would be to simply leave the question up to their Red List and let them worry about particulars. If I want to know if species X was extant in 2004, I check their database and see if its designation was something other than "extinct" for that year. They may not always be able to pinpoint the exact time of death for every species, but they do as good a job as anyone, or better.Of course, the IUCN doesn't cover all species, leaving out 1) species that have been extinct for a long time (e.g., Tyrannosaurus rex), and 2) species that haven't been published by scientists yet (e.g., Laonastes aenigmamus in lists prior to 2005). But I think in both of these cases we can consider such species to be "non-extant for the purposes of nomenclature". Long-extinct species are clearly not extant. Treating undiscovered species as non-extant has the same stabilizing benefit as requiring an extant species to be published upon. The only problem spot is the taxa that the IUCN doesn't focus on, e.g., bacteria and archaeans. But this still leaves plenty of taxa that it works just fine for.
I think I like this approach best, at least for the taxa I study (amniotes). Delegate the issue to the experts. Mylodon and Synocnus are extinct. Lipotes is critically endangered (at least as of last year). The nomenclatural problem is taken care of, and we can move on to more crucial problems, like preserving the crown groups that we have.
Labels:
Cetacea,
clade,
conservation,
definition,
evolution,
extinction,
Folivora,
mammalia,
nomenclature,
paleontology,
PhyloCode,
phylogeny,
placentalia,
systematics,
taxonomy,
zoology
Subscribe to:
Comments (Atom)