(Click to view larger version.)
28 March 2010
05 March 2010
Names on Nodes: Cutting Out the Fat
While pondering the headaches of homonymy recently, I started to ask myself, What am I doing with my life? Why am I worrying about this?
Seriously, though. I've been working on Names on Nodes on and off for about three years, and I still haven't launched it. And it's because I've been so focused on getting things like this right. (Well, that and having a day job.)
But things like this aren't part of the core functionality. The core functionality is the automated evaluation of phylogenetic definitions (encoded as MathML) within the context of phylogenetic hypotheses (modeled as directed, acyclic graphs). That part of it's been done for quite a while. So why am I wasting time on the rest?
By cutting out the entire database portion of the project, I could actually have something launched this year. Sure, it'd be nice to have a repository of taxonomic names, definitions, authorities, etc. But it's not necessary. It's phase II, not phase I. In fact, by the time I'm ready for a phase II, there will almost certainly be other services out there that already perform those things.
So, here on out, I'm going to be focusing on getting a lean, mean version of Names on Nodes up. Here's a quick summary of what you'll be able to do with it:
Seriously, though. I've been working on Names on Nodes on and off for about three years, and I still haven't launched it. And it's because I've been so focused on getting things like this right. (Well, that and having a day job.)
But things like this aren't part of the core functionality. The core functionality is the automated evaluation of phylogenetic definitions (encoded as MathML) within the context of phylogenetic hypotheses (modeled as directed, acyclic graphs). That part of it's been done for quite a while. So why am I wasting time on the rest?
By cutting out the entire database portion of the project, I could actually have something launched this year. Sure, it'd be nice to have a repository of taxonomic names, definitions, authorities, etc. But it's not necessary. It's phase II, not phase I. In fact, by the time I'm ready for a phase II, there will almost certainly be other services out there that already perform those things.
So, here on out, I'm going to be focusing on getting a lean, mean version of Names on Nodes up. Here's a quick summary of what you'll be able to do with it:
- Open bioinformatics files (NEXUS to start with, other formats like nexml going forward).
- View the phylogenies in a pretty graphical interface.
- Merge phylogenies.
- Tweak phylogenies (adding or removing parent-child relations, adding or removing taxonomic units, equating taxonomic units, etc.).
- Formulate phylogenetic definitions using a spiffy interface.
- Apply these definitions to the phylogeny.
- Save your work as MathML files.
Labels:
databases,
definition,
MathML,
Names on Nodes,
nomenclature,
phylogeny,
RIAs,
web applications
01 March 2010
The Great PhyloCode Land Run
Sometime in the near future, the PhyloCode will be enacted. For this to happen, two things need to happen concurrently:
1. The registration database (called "RegNum") must be completed and opened to the public. This is necessary because the PhyloCode requires all names to be registered electronically.
2. Phylonyms: a Companion to the PhyloCode must be published. This is a multi-authored volume that will include the earliest definitions under the PhyloCode.
Which names will be defined in Phylonyms? The original goal was to cover the most historically important names (what Alain Dubois calls "sozonyms"). However, proponents of phylogenetic nomenclature tend to be clustered in several fields (most notably vascular plant botany and vertebrate zoology—note that the code's authorship reflects this). This means certain parts of the Tree of Life (e.g., entomology) will unfortunately be underrepresented, due to lack of interest in those fields. (The alternative, having non-specialists define such names in Phylonyms, does not bear consideration.) So Phylonyms will be less about providing coverage and more about providing sturdy, well-reasoned definitions that can serve as examples.
What about all the names that it omits? What will happen to those once the PhyloCode is enacted? That will be interesting to see.
One thing I could envision is a sort of "land run". I picture it working this way. Let's consider a field, say, anthropology, where phylogenetic nomenclature has not taken much of a hold. Currently there is debate about how to use some taxonomic names related to the field. Some workers like to use the familial name "Hominidae" to refer to a large taxon, including humans and great apes. Others prefer to restrict it to the human total clade (i.e., humans and everything closer to them than to other extant taxa). Similarly, some workers use the generic name "Homo" in a broad sense to include short, small-brained species like Homo habilis, while others prefer to restrict it to the tall, large-brained clade (relegating H. habilis to another genus, e.g., Australopithecus).
Let's say there's a researcher out there named Dr. Statler, who prefers a strict usage for "Hominidae" and a broad use for "Homo". But his colleague, Dr. Waldorf, prefers a broad usage for "Hominidae". Dr. Waldorf isn't really that interested in phylogenetic nomenclature, but when he notes that "Hominidae" is not in the registration database, he sees an opportunity. He writes a quick paper defining "Hominidae" as a node-based clade: "The clade originating with the last common ancestor of humans (Homo sapiens Linnaeus 1758), Bornean orangutans (Pongo pygmaeus Linnaeus 1760), common chimpanzees (Pan troglodytes Oken 1816, originally Simia troglodytes Blumenbach 1775), and western gorillas (Gorilla gorilla Geoffroy 1852, originally Troglodytes gorilla Savage 1847)."
Dr. Statler is, of course, outraged. Not that he cares that much about phylogenetic nomenclature, but what if anthropologists do start using it? What if someone ruins another taxonomic name? His colleagues Drs. Honeydew and Beaker prefer a strict definition of "Homo"—what if they author a paper cementing that definition under the PhyloCode?
This cannot come to pass! Dr. Statler does some reading on the code and decides that a branch-based definition would work nicely for his broader usage. He defines "Homo" as, "The clade consisting of Homo sapiens Linnaeus 1758 and all organisms that share a more recent common ancestor with H. sapiens than with Australopithecus africanus Dart 1925, Paranthropus robustus Broom 1938, Zinjanthropus boisei Leakey 1959, or Australopithecus afarensis Johanson & White 1978." This sets off another anthropologist, and soon all sorts of anthropological/primatological names are being defined under the PhyloCode, as workers struggle to assert their usages.
This is not an ideal situation. It would be much nicer if a group of anthropologists were to come together, discuss the matters rationally, and arrive at an agreement which they then publish together. But it's still not a horrible situation—at least people are defining phylogenetic names and at least interest in phylogenetic nomenclature is being spread. I can't predict the future, but I feel like this sort of "land run" is bound to occur at least in some fields—and maybe that's okay.
1. The registration database (called "RegNum") must be completed and opened to the public. This is necessary because the PhyloCode requires all names to be registered electronically.
2. Phylonyms: a Companion to the PhyloCode must be published. This is a multi-authored volume that will include the earliest definitions under the PhyloCode.
Which names will be defined in Phylonyms? The original goal was to cover the most historically important names (what Alain Dubois calls "sozonyms"). However, proponents of phylogenetic nomenclature tend to be clustered in several fields (most notably vascular plant botany and vertebrate zoology—note that the code's authorship reflects this). This means certain parts of the Tree of Life (e.g., entomology) will unfortunately be underrepresented, due to lack of interest in those fields. (The alternative, having non-specialists define such names in Phylonyms, does not bear consideration.) So Phylonyms will be less about providing coverage and more about providing sturdy, well-reasoned definitions that can serve as examples.
What about all the names that it omits? What will happen to those once the PhyloCode is enacted? That will be interesting to see.
One thing I could envision is a sort of "land run". I picture it working this way. Let's consider a field, say, anthropology, where phylogenetic nomenclature has not taken much of a hold. Currently there is debate about how to use some taxonomic names related to the field. Some workers like to use the familial name "Hominidae" to refer to a large taxon, including humans and great apes. Others prefer to restrict it to the human total clade (i.e., humans and everything closer to them than to other extant taxa). Similarly, some workers use the generic name "Homo" in a broad sense to include short, small-brained species like Homo habilis, while others prefer to restrict it to the tall, large-brained clade (relegating H. habilis to another genus, e.g., Australopithecus).
Let's say there's a researcher out there named Dr. Statler, who prefers a strict usage for "Hominidae" and a broad use for "Homo". But his colleague, Dr. Waldorf, prefers a broad usage for "Hominidae". Dr. Waldorf isn't really that interested in phylogenetic nomenclature, but when he notes that "Hominidae" is not in the registration database, he sees an opportunity. He writes a quick paper defining "Hominidae" as a node-based clade: "The clade originating with the last common ancestor of humans (Homo sapiens Linnaeus 1758), Bornean orangutans (Pongo pygmaeus Linnaeus 1760), common chimpanzees (Pan troglodytes Oken 1816, originally Simia troglodytes Blumenbach 1775), and western gorillas (Gorilla gorilla Geoffroy 1852, originally Troglodytes gorilla Savage 1847)."
Dr. Statler is, of course, outraged. Not that he cares that much about phylogenetic nomenclature, but what if anthropologists do start using it? What if someone ruins another taxonomic name? His colleagues Drs. Honeydew and Beaker prefer a strict definition of "Homo"—what if they author a paper cementing that definition under the PhyloCode?
This cannot come to pass! Dr. Statler does some reading on the code and decides that a branch-based definition would work nicely for his broader usage. He defines "Homo" as, "The clade consisting of Homo sapiens Linnaeus 1758 and all organisms that share a more recent common ancestor with H. sapiens than with Australopithecus africanus Dart 1925, Paranthropus robustus Broom 1938, Zinjanthropus boisei Leakey 1959, or Australopithecus afarensis Johanson & White 1978." This sets off another anthropologist, and soon all sorts of anthropological/primatological names are being defined under the PhyloCode, as workers struggle to assert their usages.
This is not an ideal situation. It would be much nicer if a group of anthropologists were to come together, discuss the matters rationally, and arrive at an agreement which they then publish together. But it's still not a horrible situation—at least people are defining phylogenetic names and at least interest in phylogenetic nomenclature is being spread. I can't predict the future, but I feel like this sort of "land run" is bound to occur at least in some fields—and maybe that's okay.
Labels:
anthropology,
clade,
definition,
entomology,
literature,
nomenclature,
PhyloCode,
phylogeny,
primatology,
systematics,
taxonomy,
Vertebrata,
zoology
27 February 2010
One Name, One Taxon -- For One Rank Group
How Many Taxa Per Name?
A while back I pondered a seeming contradiction between the way zoological nomenclature is practiced and what the ICZN actually says. To illustrate, let's consider the case of Columbina Illiger 1811 and Columbina Spix 1825. The former is a subtribe, typified by Genus Columba, and the latter is a genus. It's possible for Columbina Illiger 1811 to include Columbina Spix 1825, although, as I understand it, they would generally be considered disjoint taxa, with Columbina Spix 1825 in another subtribe.
In several places, it seems as though the ICZN would not allow one name to refer to different taxa. The Preamble states that one of its objectives is "to ensure that the name of each taxon is unique and distinct", and Art. 52.1 states that, "When two or more taxa are distinguished from each other they must not be denoted by the same name." Logically, it would seem that Columbina Spix 1825 should be considered invalid, and that Columbina Illiger 1811 should have priority.
But this is not how the code is interpreted. There is an understanding that homonymy only occurs within rank groups (family group, genus group, species group). Since Columbina Spix 1825 is a genus-group name and Columbina Illiger 1811 is a family-group name, they can't be homonyms. (Elsewhere, a term has been coined for such apparent homonyms: "hemihomonyms".)
This understanding is implicit. Nowhere does the ICZN explicitly lay it out. The closest it gets is in Article 53, which discusses the particulars of how homonymy works. It discusses homonymy within the family group, homonymy within the genus group, and homonymy within the species group. Nowhere does it discuss homonymy between rank groups. Only by this omission does the code hint at the idea that homonymy only occurs within rank groups.
I've communicated with several taxonomists, including people involved with the ICZN, and they all seem to agree that this is the code's intent and that the wordings in the Preamble and Art. 52.1 are confusing. Hopefully a future version of the code will clarify this.
When a Code Is Not a Namespace
So, with that more or less settled, now I'm back to my original problem. In Names on Nodes, authorities (such as nomenclatural codes) are treated as namespaces, i.e., sets of distinct names. So far as I know, there is no problem in treating the other codes (including the PhyloCode) in this manner, but apparently the ICZN does not work this way. Suppose I refer to the ICZN using a URI based on its ISBN number:
urn:isbn:0853010064. What would the qualified name urn:isbn:0853010064::Columbina refer to?Here are a few ideas I've come up with.
One Code, Three Namespaces
So the ICZN doesn't function as a namespace—but it does function as three namespaces, one for each rank group. I could use each zoological rank group as a namespace. The only problem with this is that there is no standard URI to refer to each group. At least, I don't know of any—if there is one, speak up! (I suppose I could use the draft BICI standard to refer to the particular page in the code where it defines the rank group in question, but that's a bit awkward.)
Orthographic Differences
Note that Columbina Illiger 1811 is in normal font and Columbina Spix 1825 is italicized. I could use this to distinguish the names from each other, e.g.,
urn:isbn:0853010064::Columbina (the subtribe) vs. urn:isbn:0853010064::_Columbina_ (the genus). For consistency, this would have to be done to species names as well, e.g., urn:isbn:0853010064::_Columbina+passerina_This isn't the only way to do it, though. The ICZN makes a further distinction, putting family group names in all-capital letters, e.g., COLUMBINA Illiger 1811. (Although it never states this as a rule, and most publications don't follow this convention.) I could follow this convention in the qualified names, e.g.,
urn:isbn:0853010064::COLUMBINA (the subtribe) vs. urn:isbn:0853010064::Columbina (the genus). No change would be require for qualified species names, e.g., urn:isbn:0853010064::Columbina+passerina.Augmented Local Names
Another possibility is to consider the rank group to be an essential part of the name itself. This could be reflected in a qualified name by augmenting the name with a prefix, e.g.,
urn:isbn:0853010064::fam:Columbina (the subtribe) vs. urn:isbn:0853010064::gen:Columbina (the genus). To be consistent, this would have to be applied to species names as well, e.g., urn:isbn:0853010064::sp:Columbina+passerina.What About Other Names?
The ICZN has few rules to do with names above the level of the family group, and overall it doesn't govern much about them. Thus there are all kinds of examples of homonymous taxa above the rank of family group. For example, Pterodactyloidea Plieninger 1901 is a suborder which includes Pterodactyloidea Meyer 1830, a superfamily. "Decapoda" is the name of an order-group taxon in two different phyla, Arthropoda and Mollusca. Etc., etc.
I had wanted to be able to use qualified names for all zoological names, but I'm having trouble seeing how that will be possible for those ranked above the family group. I'll probably have to use the coining publications themselves as authorities, or a URI (e.g., an LSID) for each name. Rather inconvenient.
Labels:
Names on Nodes,
nomenclature,
taxonomy,
zoology
Defining Rank-Based Taxa Mathematically
Let U be the set of all individuals.
Let ranks be represented by a contiguous series of natural numbers (ℕ). Let 1 represent the lowest (finest) rank and let some natural number n represent the highest (coarsest) rank.
Let T be a sequence of n sets of type individuals (i.e., individuals represented by type specimens). Let each set in the sequence (other than the last set) be a superset of the next set, i.e., T1 ⊇ T2 ⊇ … Tn.
Let d be a metric function measuring some distance between any two individuals: d(x, y) ∈ ℝ0+ (the set of nonnegative real numbers). Note that, because it is a metric, d(x, x) = 0 and d(x, y) = d(y, x).
For each rank level r, let pr be a function mapping each member, t, of Tr to a taxon (set of individuals): pr(t) := {x ∈ U | for all s ∈ Tr, d(x, t) ≤ d(x, s)}. Let Pr be the image of pr. Then Pr is the taxonomy of rank level r.
Note that some individuals may be placed in multiple taxa of the same rank if they are equidistant between type individuals. These individuals may be considered unclassifiable for that rank. Let U′ be the set of all individuals except for those which are unclassifiable for some rank. Similarly, let P′r be Pr but with all unclassifiable individuals removed from each member taxon. P′r is a partition on U′. For any two rank levels q and r, if q < r, thenP′q is a refinement of (or equal to) P′r .
Let ranks be represented by a contiguous series of natural numbers (ℕ). Let 1 represent the lowest (finest) rank and let some natural number n represent the highest (coarsest) rank.
Let T be a sequence of n sets of type individuals (i.e., individuals represented by type specimens). Let each set in the sequence (other than the last set) be a superset of the next set, i.e., T1 ⊇ T2 ⊇ … Tn.
Let d be a metric function measuring some distance between any two individuals: d(x, y) ∈ ℝ0+ (the set of nonnegative real numbers). Note that, because it is a metric, d(x, x) = 0 and d(x, y) = d(y, x).
For each rank level r, let pr be a function mapping each member, t, of Tr to a taxon (set of individuals): pr(t) := {x ∈ U | for all s ∈ Tr, d(x, t) ≤ d(x, s)}. Let Pr be the image of pr. Then Pr is the taxonomy of rank level r.
Note that some individuals may be placed in multiple taxa of the same rank if they are equidistant between type individuals. These individuals may be considered unclassifiable for that rank. Let U′ be the set of all individuals except for those which are unclassifiable for some rank. Similarly, let P′r be Pr but with all unclassifiable individuals removed from each member taxon. P′r is a partition on U′. For any two rank levels q and r, if q < r, then
25 February 2010
Tricksy Definitions Expressed Mathematically
Just for fun, here are a few definitions of nonstandard type to go along with those in the previous post. As any practitioner of phylogenetic nomenclature knows, most definitions are node-, branch-, or apomorphy-based, but there have been a few that don't fall into these categories.
Here are Wagner's (2004) definitions of Panbiota and Biota:
Panbiota := (Clade ∘ prc∩)(Homo sapiens).
Biota := Crown(Panbiota, "extant as of or after 2004").
This is one of the few cases where it makes more sense to define the crown clade based on the total clade rather than vice versa. (Maybe the only case? Not sure.) Technically, Wagner's wording for the definition of Panbiota might be better translated as (suc∪ ∘ min ∘ prc∩)(Homo sapiens), but it works out to the same thing.
And here's Clarke's (2004) definition of Ichthyornis:
Let M := "apomorphy 2" ∩ "apomorphy 5" ∩ "apomorphy 6" ∩ "apomorphy 7" ∩ "apomorphy 8".
(These refer to apomorphies in Clarke's Ichthyornis dispar Diagnosis.)
Ichthyornithes := Clade(YPM 1450 ← Struthio camelus ∪ Tinamus major ∪ Vultur gryphus).
("YPM" refers to the Yale Peabody Museum's Vertebrate Paleontology collection. YPM 1450 is the Ichthyornis dispar holotype specimen.)
Ichthyornis := Clade((M @ YPM 1450) ∩ Ichthyornithes).
Here are Wagner's (2004) definitions of Panbiota and Biota:
Panbiota := (Clade ∘ prc∩)(Homo sapiens).
Biota := Crown(Panbiota, "extant as of or after 2004").
This is one of the few cases where it makes more sense to define the crown clade based on the total clade rather than vice versa. (Maybe the only case? Not sure.) Technically, Wagner's wording for the definition of Panbiota might be better translated as (suc∪ ∘ min ∘ prc∩)(Homo sapiens), but it works out to the same thing.
And here's Clarke's (2004) definition of Ichthyornis:
Let M := "apomorphy 2" ∩ "apomorphy 5" ∩ "apomorphy 6" ∩ "apomorphy 7" ∩ "apomorphy 8".
(These refer to apomorphies in Clarke's Ichthyornis dispar Diagnosis.)
Ichthyornithes := Clade(YPM 1450 ← Struthio camelus ∪ Tinamus major ∪ Vultur gryphus).
("YPM" refers to the Yale Peabody Museum's Vertebrate Paleontology collection. YPM 1450 is the Ichthyornis dispar holotype specimen.)
Ichthyornis := Clade((M @ YPM 1450) ∩ Ichthyornithes).
Labels:
clade,
definition,
mathematics,
MathML,
Names on Nodes,
nomenclature,
phylogeny
Names on Nodes: MathML Definitions (Version 1.1)
After posting Version 1.0 earlier this week, I had a revelation: the cladogen functions are completely unnecessary, and everything would work a lot nicer if I just tossed them. I also realized that there really was no reason I couldn't include the various relations (precedence, immediate precedence, proper precedence, etc.), just in case anyone wanted to do some seriously non-standard definitions. After some significant revisions, I present Version 1.1.
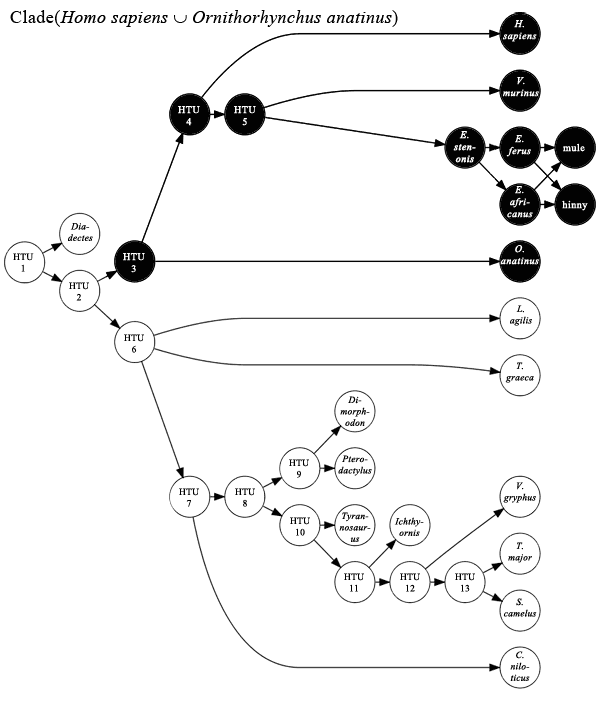
Some examples of the updated notation, using humans (Homo sapiens), platypuses (Ornithorhynchus anatinus), and Dimetrodon grandis, a stem-mammal:
Union. Homo sapiens ∪ Ornithorhynchus anatinus = all humans and all platypuses (polyphyletic taxon, also monothetic)
Exclusive Predecessors. Homo sapiens ← Ornithorhynchus anatinus = humans and all of their ancestors, except for the ancestors shared with platypuses (lineage)
Synapomorphic Predecessors. "milk glands" @ Homo sapiens = humans and all human ancestors to possess milk glands synapomorphic with those in humans (lineage)
Node-Based Clade. Clade(Homo sapiens ∪ Ornithorhynchus anatinus) = Mammalia
Branch-Based Clade (simple). Clade(Homo sapiens ← Ornithorhynchus anatinus) = "Pan-Theria"
Branch-Based Clade (multiple external specifiers). Clade(Homo sapiens ← Ornithorhynchus anatinus ∪ Dimetrodon grandis) = "Pan-Theria"
Branch-Based Clade (multiple internal specifiers). Clade(Homo sapiens ∪ Ornithorhynchus anatinus ← Dimetrodon grandis) = (unnamed clade comprised mostly of Therapsida)
Null Branch-Based Definition (multiple internal specifiers). Clade(Homo sapiens ∪ Dimetrodon grandis ← Ornithorhynchus anatinus) = ∅
Apomorphy-Based Clade. Clade("milk glands" @ Homo sapiens) = "Apo-Mammalia"
Node-Modified Crown Clade. Crown(Homo sapiens ∪ Dimetrodon grandis, "extant as of or after 2010") = Mammalia
Branch-Modified Crown Clade. Crown(Homo sapiens ← Ornithorhynchus anatinus, "extant as of or after 2010") = Theria
Apomorphy-Modified Crown Clade. Crown("milk glands" @ Homo sapiens, "extant as of or after 2010") = Mammalia
Total Clade. Total(Mammalia, "extant as of or after 2010") = Synapsida (or "Pan-Mammalia")
Some examples of the updated notation, using humans (Homo sapiens), platypuses (Ornithorhynchus anatinus), and Dimetrodon grandis, a stem-mammal:
Union. Homo sapiens ∪ Ornithorhynchus anatinus = all humans and all platypuses (polyphyletic taxon, also monothetic)
Exclusive Predecessors. Homo sapiens ← Ornithorhynchus anatinus = humans and all of their ancestors, except for the ancestors shared with platypuses (lineage)
Synapomorphic Predecessors. "milk glands" @ Homo sapiens = humans and all human ancestors to possess milk glands synapomorphic with those in humans (lineage)
Node-Based Clade. Clade(Homo sapiens ∪ Ornithorhynchus anatinus) = Mammalia
Branch-Based Clade (simple). Clade(Homo sapiens ← Ornithorhynchus anatinus) = "Pan-Theria"
Branch-Based Clade (multiple external specifiers). Clade(Homo sapiens ← Ornithorhynchus anatinus ∪ Dimetrodon grandis) = "Pan-Theria"
Branch-Based Clade (multiple internal specifiers). Clade(Homo sapiens ∪ Ornithorhynchus anatinus ← Dimetrodon grandis) = (unnamed clade comprised mostly of Therapsida)
Null Branch-Based Definition (multiple internal specifiers). Clade(Homo sapiens ∪ Dimetrodon grandis ← Ornithorhynchus anatinus) = ∅
Apomorphy-Based Clade. Clade("milk glands" @ Homo sapiens) = "Apo-Mammalia"
Node-Modified Crown Clade. Crown(Homo sapiens ∪ Dimetrodon grandis, "extant as of or after 2010") = Mammalia
Branch-Modified Crown Clade. Crown(Homo sapiens ← Ornithorhynchus anatinus, "extant as of or after 2010") = Theria
Apomorphy-Modified Crown Clade. Crown("milk glands" @ Homo sapiens, "extant as of or after 2010") = Mammalia
Total Clade. Total(Mammalia, "extant as of or after 2010") = Synapsida (or "Pan-Mammalia")

Image showing a node-based clade (Mammalia) under a given phylogenetic hypothesis. Click to enlarge. More here.
Labels:
clade,
definition,
diagrams,
mammalia,
mathematics,
MathML,
monotremata,
Names on Nodes,
nomenclature,
phylogeny,
taxonomy,
web applications,
XML,
zoology
Subscribe to:
Posts (Atom)




